
Rapid Communication
Austin J Public Health Epidemiol. 2021; 8(1): 1096.
Determination of IgG Response Profile in SARS-CoV-2 Patients Using a Multiplex Serological Assay
Brochot E1,2*, Souplet V3*, Follet P3, Ponthieu P3, Olivier C3, Even G4, Audebert C4 and Malbec R4
1Department of Virology, Amiens University Medical Center, Amiens, France
2Agents infectieux résistance et chimiothérapie Research Unit, UR4294, Jules Verne University of Picardie, France
3Innobiochips, 70 rue du Dr Yersin, 59120 Loos, France
4GD Biotech, 3595 Route de Tournai, 59501 Douai, France
*Corresponding author: Etienne Brochot, Department of Virology, Amiens University Medical Center, Amiens, France; Agents infectieux résistance et chimiothérapie Research Unit, UR4294, Jules Verne University of Picardie, France
Vianney Souplet, Innobiochips, 70 rue du Dr Yersin, 59120 Loos, France
Received: April 16, 2021; Accepted: May 11, 2021; Published: May 18, 2021
Abstract
Background: Beyond the diagnosis of SARS-CoV-2 infection, tools delivering a global picture of the patients’ humoral response may be of interest for the comprehension of the disease severity and the assessment of the patients’ protection for vaccination strategy.
Objectives: Here we use a commercial multiplex serological immunoassay CoViDiag®, based on an array of five different antigens of the virus (the Nucleocapsid, the Spike 1 and Spike 2 subunits, and the RBD and NTD domains of the Spike), to investigate the profile of the IgG humoral response for patients with recent SARS-CoV-2 infection depending on the disease severity outcome, or the time post-PCR.
Results: No cross-reaction was observed with the four other seasonal coronaviruses (100% specificity, 0/28). 100% (20/20) of the hospitalized patients PCR-positive to SARS-CoV-2 presented detectable levels of IgGs. 14 days post-PCR diagnosis, 92.3% of the patients, PCR-positive, that did not required hospitalization are presenting IgG (36/39). Interestingly for CoViDiag-positive samples, detectable levels of anti-RBD were found mainly in hospitalized patients (85%, 17/20), while the presence of anti-S1 (60.9%, 28/46) combined with the absence of anti-RBD (6.5%, 3/46) was more characteristic of nonhospitalized patients. Screening campaign group lacked both anti-S1 (18.2%, 4/22) and anti-RBD (4.5%, 1/22).
Conclusion: The CoViDiag® IgG assay could be used to evaluate patients’ immunization and improve their management.
Keywords: SARS-CoV-2; COVID-19; Serological assays; Multiplexing; IgG profile
Background
Since its first detection in Wuhan (China) in December 2019, the Severe Acute Respiratory Syndrome Coronavirus 2 (SARSCoV- 2) has rapidly spread to reach other countries worldwide as the coronavirus 2019 disease (COVID-19) became pandemic [1]. SARSCoV- 2 is spreading through human-to-human contact and can cause respiratory infections among others illness. The clinical picture is very diverse, from asymptomatic infections of healthy carriers, which will increase the disease spreading, to fever, dry cough, breathing difficulties, headache, or pneumonia which make it difficult to differentiate from other respiratory diseases such as flus or human Coronaviruses (hCoVs). Moreover, if most cases are classified as mild (no or moderate signs) in the first stage of the disease, it can rapidly evolve to more severe and critical states and even cause death.
The virions has a nucleocapsid composed by genomic RNA and phosphorylated Nucleocapsid (N) protein, which is buried inside a phospholipid bilayer and covered by the Spike proteins trimmers (S) that gives the CoVs their crown-like appearance on which their names are based. The S protein has two subunits, the Spike 1 (S1) which contains the Receptor-Binding Domain (RBD) and N-Terminal Domain (NTD) and the Spike 2 (S2) [2]. The choice of the antigenic domain is important, as it must be specific to the SARS-CoV-2 for discrimination against other hCoVs for example, and sensitive enough so infection would not be missed [3]. Most commercial serological assays have demonstrated satisfying performances in terms of diagnostic sensitivity and specificity, based on one of those main different antigenic domains [4,5]. It is now generally admitted that severe form of the disease are often associated to excessive immune response and “cytokine storms” [6]. However, kinetics of antibody response and protection efficiency remains poorly understood, especially several months after infection [4,7].
Objectives
The combination of different antigens could give a more comprehensive picture of the humoral response strength and diversity [8-10]. Thus, this study evaluates the immune profiling performances of the commercial multiplex immunoassay CoViDiag® targeting IgG antibodies against the N, S1, S1-RBD, S1-NTD, and S2 antigens (Figure 1), and its prognosis potential by investigating antibody patterns based on the time post-infection and the disease severity.
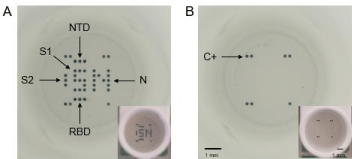
Figure 1: Full well pictures obtained with the microplate reader (SciReader®) or with a phone camera (in insert) after incubation with the CoViDiag® assay. A)
Positive sample presenting antibodies against the Nucleopcapside (N), Spike 1 (S1), N-Terminal Domain (NTD) and Receptor Binding Domain (RBD) of the Spike
protein, or Spike 2 (S2) antigens. B) Negative sample with positive control on the edges. Scale bars correspond to 1mm./div>
Material and Methods
Study design and cohort
The study was conducted at Amiens University medical Center (France). Samples were derived from de-identified excess serum
specimens as described in a previous study (Brochot et al., 2020b).
The demographic information of the 167 patients are available in
Supplementary Table 1. The study was approved by the institutional
review board of the Amiens University Medical Center (number
PI2020_843_0046, 21 April 2020).
Table 1: Diagnostic performances of the CoViDiag® and three other commercial IgG serological assays. Diagnostic sensitivity and specificity observed for different
patient groups: PCR-positive-hospitalized patients, PCR-positive-non-hospitalized patients, patients from screening campaign, and patients from control group PCRpositive
to other hCoVs.
Assay name
CoViDiag®
EuroImmune®
Diasorin®
Abbott®
Type of immunoglobulins
IgG
IgG
IgG
IgG
Antigen
N, S1, RBD, NTD,S2
S1
S1/S2
N
Patient’s group
Number of patients
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
Diagnostic Sensitivity
PCR positive
Hospitalized
20
100%
-
100%
-
100%
-
100%
-
Non-hospitalized
57
80.70%
-
77.20%
-
70.20%
-
80.70%
-
Screening Campaigns
62
35.50%
-
37.10%
-
21%
-
29%
-
hCoV control group (before 2020)
28
-
100%
-
96.40%
-
100%
-
100%
Table 1: Diagnostic performances of the CoViDiag® and three other commercial IgG serological assays. Diagnostic sensitivity and specificity observed for different
patient groups: PCR-positive-hospitalized patients, PCR-positive-non-hospitalized patients, patients from screening campaign, and patients from control group PCRpositive
to other hCoVs.
Briefly n=167 sera samples from patients PCR-positive to SARSCoV-
2 and hospitalized (n=20), non-hospitalized patients but PCRpositive
to SARS-CoV-2 (n=57), patients participating in screening
campaigns (n=62), and a control group of patients with a history of
other seasonal coronavirus infection (n=28) before 2020. Sera from
patients PCR-positive to SARS-CoV-2 were collected between 0 to 80
days post-PCR.
All samples have been tested on the CoViDiag® serological assay
and compared to the results obtained with three other IgG assays
widely used worldwide (Euroimmun®, Abbott® and Diasorin®) [4].
CoViDiag® assay and analysis
The assays have been performed according to the manufacturer
instructions. The results have been automatically delivered using
the SciReader® plate reader (Scenion GmbH) and associated analysis
software, and an algorithm combining different cut-offs for the
different antigens according to the manufacturer instructions
(Supplementary Table 1).
Data and statistical analysis
The demographic information of the 167 patients has previously
been described [4].
Diagnostic specificity was evaluated on samples PCR-negative
to SARS-CoV-2 but PCR-positive to other hCoVs. Diagnostic
sensitivity was evaluated on samples PCR-positive to SARS-CoV-2
collected between 0 to 80 days post-PCR from hospitalized or nonhospitalized
patients.
For the statistical analysis, Generalized Additive Models (GAM)
were used to calculate Odds Ratios (OR) and 95% Confidence
Interval (CI) considering positivity/negativity for CoViDiag, N,
S1, S2, NTD and RBD as the main outcomes (borderline results
have been filtered for group to group comparison) and controlling
for personal background effects (sex and age). No influence of the
delay between PCR and serology has been observed. The general significance level was set at a p-value below 0.05. All analyses were
performed using packages stats and odds ratio from the R statistical
computing program v. 3.6.1 (Date of release 07/05/2019). Specifically,
we compare the antibody response profile between patients group
and depending on the time post-PCR to test whether a significant
difference is present among different group variables.
Results
Diagnostic performances of the multiplex CoViDiag® IgG
assay
All patients hospitalized for COVID-19 with a positive
nasopharyngeal SARS-CoV-2 PCR were positive to the CoViDiag®
IgG assay (n=20/20, 100%) (Table 1). We observed than only 80.7%
of the patients with a positive SARS-CoV-2 PCR that did not require
hospitalization were positive to the CoViDiag® IgG assay (n=46/57).
The part of patients presenting IgG increases to 92.3% for samples
collected at least 14 days after a positive PCR (n=36/39). We found
35.5 % of the patients participating in the screening campaigns
positives to the CoViDiag® IgG assay (n=22/62). Using the CoViDiag®
assay, we observed that 25.8% (n=16/62) of the patients from the
screening campaign were lacking either the anti-N, anti-S1 or
anti-S2 antibodies. Similar incomplete response was observed for
31.6% (n=18/57) of the non-hospitalized patients, and 10% of the hospitalized ones (n=2/20). Among the 57 non-hospitalized patients,
five presented only anti-S2 IgG (see supplementary Table 1). Among
the 62 screening campaign patients, four presented only anti-S2, two
only anti-N, and one only anti-NTD antibodies, highlighting the
interest of targeting a wide scope of antibodies especially in the light
form of the disease. There was no cross reactivity with the samples
from patients PCR positive to other seasonal coronaviruses (OC43,
HKU1, NL63, 229E), collected between day 7 and day 1153 post PCR
(100 % diagnostic specificity, n=0/28).
Profile of the IgG antibody responses depending on the
disease severity
For the patients presenting a positive IgG response to CoViDiag®,
we find different profile of the immune response between the
different patient groups (Figure 2). 95% (n=19/20) of the patients
hospitalized presented anti-S1 IgG against 60.9% (n=28/46) of the
patients non-hospitalized and 18.2% (n=4/22) of the patients from
the screening campaign. Furthermore 85% (n=17/20) of the patients
hospitalized presented anti-RBD IgG against 6.5% (n=3/46) of the
patients non-hospitalized. The comparison of odds ratio for each
antigen (Supplementary Table 2) confirmed that the presence of
anti-RBD antibodies is the best marker for the chance of being in the
hospitalized group versus non-hospitalized group (OR: 4.508, CI:
4.332-4.693, p-value: 7.34e-13) or screening campaign group (OR:
4.665, CI: 4.739-4.592, p-value: 2.48e-12). The presence of anti-S1
antibodies is the best marker for the chance of being in the nonhospitalized
group versus screening campaign group (OR: 1.901, CI:
2.044-1.767, p-value: 0.002).
Figure 2: IgG profile of CoViDiag-positive patients: percentage of patients,
positives to the CoViDiag assay, and with detectable levels of anti-N, anti-S1,
anti-NTD, anti-RBD, and anti-S2 antibodies, depending on the disease
outcome severity.
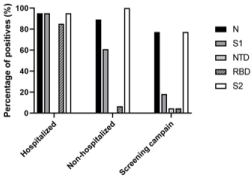
Figure 2: IgG profile of CoViDiag-positive patients: percentage of patients,
positives to the CoViDiag assay, and with detectable levels of anti-N, anti-S1,
anti-NTD, anti-RBD, and anti-S2 antibodies, depending on the disease
outcome severity.
Profile of the IgG antibody responses depending on the
time post-PCR
For the two groups of patients PCR positive to SARS-CoV-2,
we investigated the profile of the IgG antibody responses depending
on the delay between PCR and serology. Independently of the
period of collection between 0 and 80 days post-PCR the majority
of hospitalized patients presented detectable levels of anti-N, anti-S1,
anti-RBD and anti-S2 IgG antibodies, but no anti-NTD antibodies
(Figure 3A). However, for the non-hospitalized patients, the immune
response appeared weaker, allowing to follow the IgG antibody
different kinetics (Figure 3B). The number of patients with anti-N,
anti-S1, anti-RBD and anti-S2 IgG antibodies, increased until 45 days
post-PCR, before starting to drop, especially for anti-N IgG antibodies
(Δ= -27.5% between 31-45 and >45 days post-PCR). Furthermore in
the 14 days following the PCR, the anti-N and anti-S2 are the main
detected IgG antibodies (44.4% anti-N positives and 61.1% anti-S2
positives) while the anti-S1 IgG antibodies are generally detected
latter (50% anti-S1 positives between 15-30 days).
Figure 3: Evolution of the IgG profile for hospitalized; A) and non-hospitalized patients; B) Percentage of patients with detectable levels of anti-N, anti-S1, anti-NTD,
anti-RBD, and anti-S2 antibodies, depending on the delay between PCR and serology.
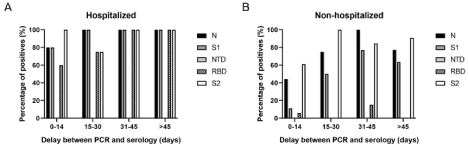
Figure 3: Evolution of the IgG profile for hospitalized; A) and non-hospitalized patients; B) Percentage of patients with detectable levels of anti-N, anti-S1, anti-NTD,
anti-RBD, and anti-S2 antibodies, depending on the delay between PCR and serology.
Discussion
For routine diagnosis use, commercial serological assays must be
evaluated in regard to their ability to detect early and weak infections.
Several commercial assays have shown good performances focusing
on the detection of total antibodies (IgG, IgM and IgA). However, as
early diagnosis results are already delivered by PCR assays, serological
assays detecting IgG seem more appropriate for the evaluation of an
efficient and long lasting protection of the patients. Interestingly, the
detection of antibodies against larger specter of antigens can also
increase the diagnostic sensitivity, especially for generally weaker
immune response of asymptomatic and mild forms. With diagnostic
performances equivalent to other IgG commercial serological assays,
the CoViDiag® multiplex assay gives a more comprehensive picture of the IgG humoral response. This study investigates the profile of anti-
SARS-CoV-2 different antibodies. We observed different pattern of
IgG profiles between severe (hospitalized patients and PCR positives),
mild (non-hospitalized patients and PCR positives), or asymptomatic
(patients from the screening campaigns) form of the disease. On
samples more than 45 days post-PCR, the percentage of different
IgG positive results tends to decrease or remain constant for the mild
and more severe form of the diseases, respectively. Furthermore, a
lot of interrogations have been raised lately regarding the vaccination
protocol for previously infected patients. As most vaccines are based
on the RBD part of the S1 protein, multiplex serology has the potential
to differentiate between infection and vaccination, and between
variants, with a single assay. Future epidemical study on a larger
panel of samples (especially extended to the population with mild or
asymptomatic form of the disease), combining the multiplex assay
with machine learning can be a convenient tool to investigate the
kinetics and mechanisms of the immune response and contribute to
the development of long lasting and efficient strategy of vaccination.
Funding
Laboratory’s own resources
Declaration of Competing Interest
Authors Rémi Malbec, Gaël Even and Christophe Audebert
are employees of GD Biotech, while Pauline Ponthieu, Pauline
Follet, Vianney Souplet and Christophe Olivier are employees of
Innobiochips, providing the CoViDiag® assay kits for this study.
References
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia
outbreak associated with a new coronavirus of probable bat origin. Nature.
2020; 579: 270-273.
- Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and
immune responses. Journal of Medical Virology. 2020; 92: 424-432.
- Brochot E, Demey B, Touze A, Belouzard S, Dubuisson J, Schmit J-L, et
al. Anti-Spike, anti-Nucleocapsid and neutralizing antibodies in SARS-CoV-2
hospitalized patients and asymptomatic carriers (Infectious Diseases (except
HIV/AIDS)). 2020a.
- Brochot E, Demey B, Handala L, François C, Duverlie G and Castelain S.
Comparison of different serological assays for SARS-CoV-2 in real life. J Clin
Virol. 2020b; 130: 104569.
- Tuaillon E. Detection of SARS-CoV-2 antibodies using commercial assays
and seroconversion patterns in hospitalized patients. MedRxiv. 2020.
- Krajewski R, Golebiowska J, Makuch S, Mazur G and Agrawal S. Update on
serologic testing in COVID–19. Clin Chim Acta. 2020; 510: 746-750.
- Ota M. Will we see protection or reinfection in COVID-19? Nature Reviews
Immunology. 2020; 20: 351-351.
- de Assis RR, Jain A, Nakajima R, Jasinskas A, Felgner J, Obiero JM, et al.
Analysis of SARS-CoV-2 Antibodies in COVID-19 Convalescent Blood using
a Coronavirus Antigen Microarray (Immunology). 2020.
- Coste AT, Jaton K, Papadimitriou-Olivgeris M, Greub G and Croxatto A.
Comparison of SARS-CoV-2 serological tests with different antigen targets
(Infectious Diseases (except HIV/AIDS)). 2020.
- Lynch KL, Whitman JD, Lacanienta NP, Beckerdite EW, Kastner SA, Shy
BR, et al. Magnitude and kinetics of anti-SARS-CoV-2 antibody responses
and their relationship to disease severity (Infectious Diseases (except HIV/
AIDS)). 2020.