
Research Article
Austin J Microbiol. 2019; 5(1): 1025.
In-Vitro and In-Vivo Pathogenicity of Mar, Biofilm Forming Non-Cholera Vibrios (NCV) From Asian Tiger Shrimp (Penaeus monodon): Implications for Food Safety and Sanitation
Nandita M, Sarita G Bhat* and Harshan A
Department of Biotechnology, Cochin University of Science and Technology, India
*Corresponding author: Sarita G Bhat, Department of Biotechnology, Cochin University of Science and Technology, Kochi-682022, Kerala, India
Received: May 15, 2019; Accepted: June 04, 2019; Published: June 11, 2019
Abstract
Contamination of aquatic environment with antibiotic resistant bacteria is a multi-factorial global threat with deleterious effects on human and animal health. In this preliminary study, we examined the in-vitro susceptibility of 14 antibiotics against 4 biofilm forming non-cholera Vibrios, (Vibrio alginolyticus and Vibrio parahemolyticus) isolated from fresh and healthy Asian Tiger Shrimp (Penaeus monodon), sourced from retail fish markets in Cochin during a three-month period. Biofilm forming capacities were evaluated by qualitative and quantitative assays as well as by characterization of biofilms under different food related stress conditions. Growth temperature and NaCl concentration influenced biofilm formation profoundly. Antibiotic resistance/susceptibility profiles of 4 biofilm formers assessed by Kirby-Bauer disc diffusion method, showed resistance to 13 of the 14 antibiotics tested. The pathogenicity profile of the isolates was elucidated by in-vitro assays like exo-enzyme profiling, auto-aggregation and surface hydrophobicity. In addition, a Caenorhabditis elegans based in-vivo pathogenicity testing by survival score analysis was also included. All these findings reveal that shrimp or related seafood harbors antibiotic resistant, biofilm forming Vibrios, indicating high-risk of food-related illnesses in humans. Further understanding of these processes will provide novel insights into the therapeutics and prevention of biofilm-related Vibrio infections in the aquaculture/seafood industry.
Keywords: Vibrios; Shrimp; Antibiotic resistance; Pathogenicity; Biofilms; Food contamination; C.elegans
Introduction
Bacterial adherence to food products or contact surfaces is a significant source of contamination, causing hygiene/health issues as well as economic losses in the seafood industry. Biofilms being a predominant and successful mode of microbial life, can implicitly develop on natural as well as man-made [1]. These heterogeneous microbial communities enclosed in a self-synthesized layer of complex polysaccharides, proteins, lipids and extracellular DNA, collectively called the extracellular polymeric substance or EPS [2,3], can colonize different sea foods like cockles, shrimp, crabs etc raising food safety concerns on a global scale.
Farmed shrimp are administered antibiotics like gentamicin, sulphonamides, tetracyclines, chloramphenicol, trimethoprim, fluoroquinolones etc., on a regular basis to prevent or treat bacterial diseases [4]. Even as tetracycline is recommended in shrimp farming [5], the last decade saw several reports on the acquisition of tetracycline resistance via plasmids or other mobile genetic elements [6]. Antibiotic resistant halophilic `Non-Cholera Vibrios’ (NCV’s) associated with shrimp farming in India have been documented [7,8]. However, they do not satisfactory correlate (if any) biofilm formation, antibiotic resistance and virulence potential of Vibrios. The dangerous spread of biofilm related bacterial infections augmented the demand to study ‘host pathogen interactions’ using Caenorhabditis elegans as a resourceful model. This pilot study was therefore undertaken to evaluate antibiotic susceptibility and virulence potential of biofilm forming NCV’s isolated from edible shrimp (Penaeus monodon).
Materials and Methods
Sampling
10 shrimp sampling, from four retail fish markets in Cochin, South India over a three month period from January to March 2017 were done. The head and tail were removed and the gut homogenized with 0.85% saline; 25g of homogenate added to 225mL of Alkaline Peptone Water (APW) pH 8.6, and incubated at 370 C for 24 hours. Two loopful of culture from pellicle of each flask with APW were plated on Thiosulfate Citrate Bile Salts Sucrose (TCBS) agar plates (HiMedia, Mumbai, India) and incubated at 370C for 24 hours. Green and yellow colonies (3-5mm diameter) were randomly selected and inoculated onto Vibrio-specific agars such as Vibrio Alginolyticus agar (VAL) [9] and Vibrio Parahaemolyticus Sucrose Agar (VPSA) (HiMedia, Mumbai, India) and incubated overnight for confirmation.
Biochemical characterization of bacterial strains
Phenotypic characterization of the isolates was as outlined in Bergey’s Manual of Systematic Bacteriology, by using KB007 Hi- Vibrio™ Identification Kit.
Molecular confirmation of Vibrio species
Genomic DNA was isolated and purified and a portion of the 16S rDNA was amplified using a universal primer pair for 16S rDNA. The sequences for the primer pair is as follows:-Forward primer -5’ AGAGTTTGATCCTGGCTCAG 3’. Reverse primer - 5’ACGGCTACCTTGTTACGACTT 3’.The identity of the sequences was determined by comparing the 16S rDNA sequence with the sequences available in the NCBI nucleotide databases using BLAST (Basic Local Alignment Search Tool) algorithm [10]. A phylogenetic tree was constructed by comparing the present isolates with Vibrio phylogeny retrieved from GenBank database by the Neighbor- Joining method [11] using the MEGA 4 software [12]. The obtained sequences were aligned and submitted in GenBank.
Screening of biofilm formers among Vibrio isolates
Qualitative assessment of biofilms: Congo Red Agar method [13] and Tube adherence method [14] were used with modifications for qualitative assessment of biofilms methods. The bacterial strains were incubated overnight on CRA plates and observed for the presence of rough black crystalline colonies which are indicative of biofilm production. For tube adherence test, strains were grown in nutrient broth under shaking conditions to allow biofilm formation and then transferred to glass test tubes, incubated without shaking for 18 hours at 28°C; the culture broth was discarded and the biofilm at the interface between the air and medium was visualized using 1% Crystal Violet (CV). BTSD2 (Bacillus licheniformis, accession no: KF573745) [15] was used as positive control, while un-inoculated Trypticase soy broth medium was used as a negative control for the biofilm assays.
Quantitative assessment of biofilms: The quantitative estimation of biofilm was performed by microtiter plate method [16,17]. Briefly, the overnight cultures of Vibrios were diluted (1:10 dilution) and 20 μl of the diluted broth was added to 300 μl of TSB in 96-well flatbottomed microtiter plate. After overnight incubation, the contents of each well were aspirated with PBS buffer and vigorously shaken in order to remove any non-adherent bacteria. The remaining bacteria were fixed with methanol for 15 minutes and later stained for 5 minutes with 1% CV solution. Excess stain was rinsed off and the biofilms, upon drying were extracted with 33% (v/v) glacial acetic acid per well. All tests were repeated thrice independently, and statistically analyzed.
Investigation of Vibrio biofilms under different stress conditions
Influence of incubation temperature on biofilm formation was investigated at 4°C, 280C and 370C for 24, 48 and 72 hours under static conditions using micro-titre plate method.
Influence of static and dynamic (shaking) conditions on biofilm formation - Dynamic conditions was achieved by incubating on a horizontal shaker at 150 rpm. The microtitre plates were incubated for 24, 48 and 72 hours at 280C and 370C under static and dynamic conditions
Influence of NaCl on biofilm formation used 1%, 3%, 5% 8% and 10% concentrations of NaCl in TSB for 24 hours under static conditions.
Antibiotic susceptibility testing
Antibiotic sensitivity of Vibrio isolates were tested on Mueller– Hinton (MH) agar in accordance with the Kirby- Bauer method [18], with 14 antibiotics (HiMedia, Mumbai, India) belonging to different classes, namely ampicillin (10μg/disc), azithromycin (15μg/ disc), carbenicillin (100μg/disc ), cefixime (5μg/disc), cefuroxime (30μg/disc), chloramphenicol (30μg/disc), co-trimoxazole (25μg/ disc), gentamicin (10μg/disc), nalidixic acid (30μg/disc), rifampicin (5μg/disc), streptomycin (10μg/disc), tetracycline (30μg/disc), trimethoprim (5μg/disc) and vancomycin (30μg/disc). Fresh cultures were inoculated into Luria -Bertani broth and incubated until Optical density equaled MacFarland 0.5, plated on Mueller–Hinton agar and antibiotic discs were placed. Upon incubation at 37oC for 18–20 hours, growth inhibition around discs was measured and the results were interpreted as per the manufacturers’ instructions.
In vitro pathogenicity assays for Vibrios
(a) Exoenzyme profiling was done by incorporating aesculin, starch and tributyrin into the basal medium and the plates were observed to determine the exo-enzyme production [19].
(b) Hemolytic assay-The test organisms were spot inoculated on blood agar plates, incubated at 370C for 24 hours and alpha, beta or gamma hemolysis were categorized based on the lytic zones produced [20].
(c) Auto aggregation assay and Suicide Phenomenon - Auto aggregation assay was performed according to Kos et al. [21] with modifications. Bacteria were grown for 18 hours at 370C in nutrient broth with 1% NaCl, harvested by centrifugation at 5000g for 15min, washed twice and resuspended in Phosphate Buffered Saline (PBS) to get approximately 108 CFU ml-1. Cell suspension (4mL) was vortexed for 10 sec and auto aggregation was determined during 5 hours of incubation at room temperature. Every hour 0.1mL from the upper suspension was transferred to another tube with 3.9mL of PBS and the absorbance (A) was measured at 600nm. Auto aggregation percentage is expressed as 1-(At/A0) x 100, where At represents the absorbance at time t =1, 2, 3, 4 or 5 hours and A0 the absorbance at t = 0.
Nutrient Broth with 0.5% glucose was inoculated and incubated at 37°C for 24 hours to determine suicide phenomenon. Strains showing the suicide phenomenon spontaneously pelleted, while those lacking this characteristic showed uniform broth turbidity [22].
(d) Autoagglutination & Precipitation after boiling -.Vibrio strains were inoculated into Brain Heart Infusion Broth (BHIB) and incubated at 28°C for 18 hours (static conditions) and observed for Self-Pelleting (SP). Absence of growth in the broth phase and appearance of large aggregates as a button in the bottom of the tube is the clear indication of self-pelleting. Such strains were designated as SP+. Later, the tubes were vortexed for 30 seconds and split into two equal fractions. One aliquot was held at room temperature for 1 hour, and the other one was placed in a boiling-water bath for the same period of time. Upon the end of incubation, the tubes were cooled and compared with unheated controls. Strains which exhibited a reduction in the turbidity after heat treatment (compared with unheated controls) were considered positive for Precipitation After Boiling (PAB+.). The Relative Degree of Precipitation (RDP) was calculated by measuring the absorbance at 540 nm according to the following formula: RDP = A540 (untreated) - A540 (heated) [23].
(e) Surface hydrophobicity - Cell surface hydrophobicity of bacteria was analyzed by determining microbial/bacterial adhesion to hydrocarbons (MATH or BATH) as per Rosenberg et al. [24]. Overnight bacterial cultures were centrifuged and the pellets were washed twice with PBS and resuspended in PBS (pH 7.4) to get OD600=0.1. 0.5 ml p-xylene was added to 1.2 ml aliquots. The tubes were incubated at 30oC for 10 mins, vortexed, allowed to stand at RT and the lower aqueous phase was removed and measured at OD600. The results were expressed as the percentage decrease in absorbance (OD600) of the lower aqueous phase compared with OD600 of the initial cell suspension.
In-vivo pathogenicity assays using C.elegans
(a)Nematode, general maintenance and reagents- The nematode C. elegans Wild Type N2 was propagated on Nematode Growth Medium (NGM agar) in 6 cm diameter plates and fed with Escherichia coli OP50 grown in Luria Bertani broth as per Brenner [25].
(b) Solid killing assay with C. elegans- Evaluation of C. elegans life span feeding on the experimental strains was carried out as per Aballay et al. [26]. Individual bacteria were inoculated into 5 mL of LB and grown at 28°C until it reached 0.5 OD600.10 μL was spread on NGM plates and incubated overnight. Approximately 20-25 age-synchronized (adult bleaching) L4 stage hermaphrodites were transferred from a lawn of E. coli OP50 to a lawn of the test organism, and incubated at 20°C for 24 hours. The worms were then seeded onto plates containing OP50 and scored for dead nematodes at 24 h intervals for a period of 10 days with a dissecting stereomicroscope (Labomed, CZM6) for viability.
(c)Pharyngeal pumping assay- The control and infected worms were placed on NGM plates seeded with OP50 and Vibrios respectively, and pharyngeal pumping was observed using a stereomicroscope for 30 consecutive seconds [27].
Statistical analysis
All experiments were repeated thrice and the experimental data points were plotted using GraphPad Prism (Version 6.0, CA, USA ). Data was expressed as Mean values ± Standard Errors of the Mean (SEM). The time taken for 50% of the nematodes to die (time to death 50, TD50) was calculated using the equation: Y = Bottom + (Top – Bottom)/ (1 + 10^ ((LogEC50 –X)*Hill Slope)), where X is the logarithm of days and Y is the average of dead worms.
Statistical evaluations of survival analysis between Control OP50 and pathogen groups were performed by one-way ANOVA followed by Student–Newman–Keul’s test. A p-value of less than 0.05 was considered to be significant.
Results
Isolation and biochemical identification of bacterial isolates
The shrimp gut homogenate plated on TCBS agar gave green/ yellow colonies, which were picked, and streaked on Vibrio-specific agars; three isolates (BTSV1,BTSV2 and BTMV5) appeared greenish yellow on VAL agar and one isolate (BTSV4) appeared bluish green on VPSA were considered presumptive for V. alginolyticus and V. parahaemolyticus respectively (Figure 1). The 4 isolates were biochemically characterized and identified using KB007 Hi-Vibrio™ Identification Kit (Table 1).
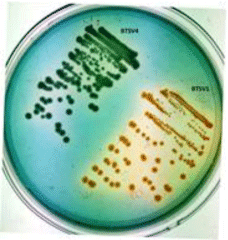
Figure 1: Bluish green colony of V. parahaemolyticus (BTSV4) and yellow
colony of V.alginolyticus (BTSV1) in VPSA.
Biochemical Tests
SV1
SV2
SV4
MV5
Gram staining
Gram negative rods
Gram negative rods
Gram negative rods
Gram negative rods
Oxidase Test
positive
positive
positive
positive
MOF Test
A/A
A/A
A/A
A/A
Voges Proskauer’s
_
_
_
_
Arginine Utilization
_
_
_
_
Salt tolerance (1%)
+
+
+
+
ONPG
_
+
_
_
Citrate Utilization
_
_
_
_
Ornithine Utilization
+
_
+
_
Mannitol
+
+
+
+
Arabinose
_
+
+
+
Sucrose
+
+
_
_
Glucose
+
+
+
+
Salicin
_
_
_
_
Cellobiose
_
_
_
_
+ = Positive; - = Negative; A/A= Acidic Slant/Acidic Butt (yellow)
Table 1: Biochemical Identification of Isolates.
Molecular confirmation of Vibrio isolates
PCR based 16S rDNA amplification (Figure 2a) and sequence analysis confirmed the molecular identity of Vibrios - three strains of V. alginolyticus (BTSV1, BTSV2, BTMV5) and one of V. parahaemolyticus (BTSV4). The sequences were submitted in GenBank and the accession numbers KY824726 (BTSV1), KY824727 (BTSV2), KY824729 (BTSV4), and KY824730 (BTMV5) obtained.
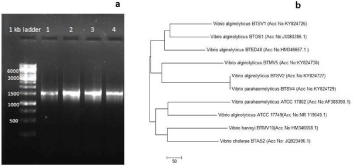
Figure 2: (a) Agarose gel showing 16S rDNA gene amplification: lane
1: BTSV1, lane 2: BTSV2, lane 3: BTSV4, lane 4 BTMV5 2(b) Showing
phylogenetic analysis of the Vibrio isolates.
V. alginolyticus strain BTSV2 and V. parahaemolyticus strain BTSV4 claded together which indicated of their genetic relatedness. The other two strains of V.alginolyticus (BTSV1 and BTMV5 existed as a separate clade. However, the isolates showed a closer similarity to two V.alginolyticus strains (BTOS1 and BTED48) retrieved from GenBank database using BLAST analysis.
Qualitative biofilm formation assays
Congo red agar method helped to differentiate strong, moderate and weak biofilm producers. Strain BTSV1 & BTSV4 produced black crystalline colonies whereas BTSV2 & BTMV5 showed smooth red coloured ones. According to the intensity of color, the isolates were categorized as strong (BTSV1 & BTSV4), moderate (BTSV2, and BTMV5) (Figure 3a).
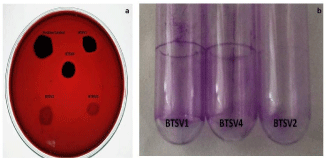
Figure 3: (a) Congo red agar plate with biofilm producers. 3(b) Comparative
view of stained tubes showing biofilm attachment- A & B-highly adherent
strains., C-weakly adherent strain.
Tube adherence method: A thick visible film lined the wall and bottom of the tube with strains BTSV1 & BTSV4, indicative of strong adherence while those with strains BTSV2 & BTMV5 had less visible film formation, suggestive of their weakly adherence to glass materials (Figure 3b).
Quantitative investigation of Vibrio biofilms under different stress conditions
Incubation temperature influenced biofilm formation: At 4 °C due to reduced growth, the biofilm formation was less evident for all strains tested. In contrast to the lower temperature, biofilm was much prominent at 280C and 370C, and the biofilm architecture was sustained up to 72 hours for all cases. BTSV2 was the strongest biofilm producer at 280C and BTSV1 at 370C (Figure 4a and 4b). The biofilm formation increased with increased incubation period except for BTSV2 and BTMV5.
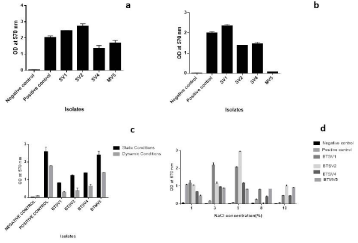
Figure 4: Influence of temperature on biofilm activity upon 72 hours of incubation (a) at 280c (b) 370c (c) Biofilm activity under static and dynamic conditions. (d)
Influence of NaCl on biofilm formation. (Negative control-TSB and Positive control-BTSD2).
Influence of static and dynamic (shaking) conditions on biofilm formation: All isolates produced biofilm in a dynamic environment. That said, the rate of biofilm formation was less than that in the static environment. It was also observed that differences in temperature (280C and 370C) and an increase in incubation time did not impact biofilm formation under dynamic conditions. Hence, biofilm assay at 370C for 24 hours under both conditions is depicted in Figure 4c.
Influence of NaCl on biofilm formation: Biofilm development at 5% NaCl was highest in the microtitre plate (Figure 4d).
Antibiotic susceptibility testing
Antibiogram showed that all four Vibrios were multiple antibiotic resistant bacteria with MAR index > 0.2 (Table 2).
Sl No
Isolates
A
B
MAR Index
(A/B)
1
BTSV1
9
14
0.62
2
BTSV2
6
14
0.42
3
BTSV4
5
14
0.35
4
BTMV5
5
14
0.35
A= No of Resistant Antibiotics; B = Total No of Antibiotics Tested
Table 2: MAR Index of Vibrios.
The isolates were characterized by resistance to 13 among 14 antibiotics tested including third generation Cephalosporins such as Cefixime. However, none were resistance to Co-trimoxazole (Figure 5).
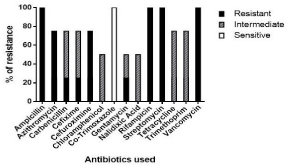
Figure 5 Antibiotic profiling of Vibrio isolates.
In- vitro assays for pathogenicity
Exoenzyme production: More than one hydrolytic enzyme was produced by the isolates which was indicative of their ability to digest nutrients present in food material. The isolates also exhibited a distinctive alpha hemolysis (partial hemolysis) pattern on blood agar plates. The results were summarized in the table below (Table 3) and (Figure 6).

Figure 6: Exoenzymes produced by Vibrios (a) Starch hydrolysis (b) Lipid hydrolysis (c) Aesculin hydrolysis (d) Hemolytic assay.
Isolates
Starch
hydrolysis
Lipid
hydrolysis
Aesculin hydrolysis
Hemolytic assay
BTSV1
+
+
+
Alpha hemolysis
BTSV2
+
+
+
Alpha hemolysis
BTSV4
+
+
+
Alpha hemolysis
BTMV5
+
+
+
Alpha hemolysis
+ = Positive; - = Negative
Table 3: Showing the enzyme profiling of Vibrios.
Auto aggregation assay: Aggregation increased with the increase in incubation time. Auto aggregating phenotype was noted in all isolates, with highest of 90% by BTSV1 (Figure 7a).
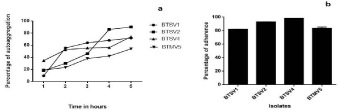
Figure 7: (a) Auto aggregation pattern of isolates (b) Cell surface hydrophobicity of isolates.
Suicide Phenomenon, auto agglutination & precipitation after boiling and Surface hydrophobicity
Thick pellets were observed for the isolates SV1 and SV2 at the bottom of the tubes upon 18 hours of incubation under static conditions, indicating that the isolates were positively suicidal (SP+) where as others were non-suicidal (SP-) strains (SV4 and MV5). The isolates were positive for auto agglutination test and precipitation after boiling (PAB+).
Percentage of hydrophobicity values less than 20 are considered as weakly hydrophobic. The isolates tested were all strongly hydrophobic on showing adherence to xylene, with BTSV4 maxing at 98.28% (Figure 7b).
Survival score analysis by solid killing assay of C. elegans
Since the Vibrios exhibited strong biofilm formation under different stress conditions, a C. elegans nematode infection model was used to evaluate their in-vivo pathogenicity. Wild-type C. elegans (N2) remained viable and healthy on the plates fed with its standard laboratory food i.e. nonpathogenic Escherichia coli strain OP50. Dead nematodes appeared as long, immobile rods, which failed to respond to external stimuli like a gentle tap with a worm loop. The TD50 value for OP50 was calculated as 12.95 ± 0.473 days. By contrast, nematodes exposed to test Vibrios exhibited significantly (P‹0.05) reduced lifespan with abnormal phenotypic behavior like reduction in pharyngeal pumping, protruding vulva and internal hatching (Figure 8).

Figure 8: (a) Healthy live C.elegans in OP50 (b) Dead nematode in OP50 (long and immotile) (c) Protruding Vulva formation (d) Internal hatching. Scale bar
represents 100 μm.
The TD50 values of the isolates BTSV1, BTSV2, BTSV4 and BTMV5 were 5.16 ±0.809 days, 4.43 ± 0.410 days, 3.57 ± 0.460 days and 5.60 ± 0.443 days respectively. The strain BTSV4 exhibited lowest of the TD50 values. Vibrios required approximately 10 days for complete killing of the nematode (Figure 9a).
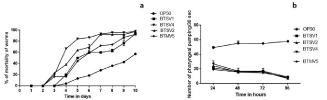
Figure 9: (a) Survival score analysis of C.elegans (b) Pharyngeal pumping assay.
Pharyngeal pumping assay
The pharyngeal pumping rate was monitored for 96 hours (four days) to assess nematode food preference. The pumping rate of the worms fed with E. coli OP50 was found to be significantly (p‹0.05) higher than pathogen fed animals and remained stable up to 4 days. The normal grazing behavior of the nematodes was affected by their exposure to Vibrios and their pumping rates notably reduced after 96 hours (Figure 9b). Worms exposed to V. parahaemolyticus (BTSV4) recorded the lowest pumping when compared to V.alginolyticuss strains mentioned in the study.
Discussion
In biofilms, bacterial aggregates are enclosed in a self-produced polysaccharide matrix attached to each other and to a biological/nonbiological surface [28,29]. Foodborne illnesses due to biofilm formers are difficult to abolish as they may be 100 times more resistant to antimicrobials compared to planktonic cells. Although virulence has been directly related to multidrug resistance, the mechanism of virulence, antibiotic resistance, and biofilm formation still remains elusive. The limited documentation in the international literature on the emergence of drug-resistant biofilm formers in market fresh shrimp was the trigger to survey these aspects. This study will therefore provide information on the existence of strong biofilm forming noncholera Vibrios isolated from fresh and healthy shrimps available at local fish markets in Cochin, South India. In addition, the antibiotic resistance profiling of these biofilm producers indicates copious use of chemicals/drugs in shrimp farming, threatening human health and safety.
Halophilic ‘Non-Cholera Vibrios’ (NCVs) are natural inhabitants of the aquatic ecosystems, chiefly associated with bacterial infections affecting all species of cultured shrimps. V. alginolyticus and V.parahaemolyticus have been etiologically associated with mass mortality of cultured shrimps and seafood-associated bacterial gastroenteritis throughout the world [30-32]. The key factor enabling Vibrios to adapt to environmental stress and transmission is the ability to form matrix-enclosed communities called biofilms [33]. With this ability to adhere to abiotic and biotic surfaces allowing their persistence and survival under aquaculture settings, the resultant Vibrios is may cause huge economic losses in the seafood industry [34]. Glass and polystyrene materials in aquaculture installations can be colonized by Vibrios as an initial step towards host colonization and thereafter to mariculture animals and seafood consumers. The isolates in this study could attach to glass and polystyrene materials at varying degree.
Studies investigating biofilm detection are performed under static conditions, though there are few reports of biofilms on surfaces under dynamic conditions. All strains in this study remained biofilm formers under both these conditions. An interpretation of this phenomenon remains unclear at this moment. The fact that shaking improves oxygenation and favorable conditions for growth of many bacteria can be attributed to this finding. It is our understanding from this study that biofilm forming bacteria can emerge as potent contaminants in water recirculation systems of various aquaculture settings, posing serious health problems in reared animals.
Nutrient availability and salinity influence biofilms by Vibrios [35]. A recent study by Han et al. [36] reported decreased biofilm formation at 4-100C, while, temperature increase enhanced biofilm formation, virulence, and quorum sensing of V. parahaemolyticus on seafood (crab, shrimp) and contact surfaces. This study too corroborates the aforesaid facts that Vibrio biofilms tend to increase with the increase in temperature and NaCl concentration. That said, the species diversity and their gene regulatory mechanisms may also play a major share in bacterial adherence.
The compact nature of biofilms contributed by matrix polymers, makes bacteria inaccessible to natural and artificial agents, and hence the ability to penetrate and destroy. Increased antibiotic resistance is a trait associated with biofilm bacteria and well elucidated in Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia), and in Vibrio cholerae. However, a correlation of antibiotic resistance and biofilm formation is still unclear in the case of non-cholera Vibrios [37].
Until recently, Vibrios were considered susceptible to many antibiotics commonly used in aquaculture. However, the past decade saw the emergence of multiple antibiotic resistant Vibrios. Resistance of V. harveyi strains to 20 different antibiotics was reported from India.13 among 14 of the antibiotics tested in our study were commonly used as prophylactic agents in shrimp farming. Among the antibiotics, the resistance to Co-trimoxazole was absent. This is in agreement with the antimicrobial pattern of luminous bacteria from shrimp farms of West Bengal studied by Sengupta et al. [38] who reported Vibrios resistant to β-lactam antibiotics and tetracycline. Only one previous study reported prevalence of antibiotic-resistant Vibrios from fish markets of Cochin [39] with V. parahaemolyticus strains resistant to ampicillin, streptomycin, and carbenicillin, but 100% susceptibility to nalidixic acid and tetracycline. They also reported 70% of isolates susceptible to trimethoprim, chloramphenicol, and gentamicin. Our study differs by indicating an alarming increase of resistance among Vibrios to a broader class of antibiotics within a short span of five years. The resistance exhibited by our isolates towards third generation cephalosporins will definetly raise the question of understanding antibiotic resistant profiles and risk factors for each pathogen.
Among the 11 species placed under ‘Harveyi Clade’ (Vibrionaceae), V. alginolyticus and V. parahaemolyticus, besides being major pathogens of marine shrimp, fish, and molluscs, are used as model organisms in biofilm studies [40], quorum sensing and multi-chromosomal genome organization [41]. The infectious life cycle of any pathogen involves different stages such as gaining entry, establishment and multiplication, avoidance of host defenses, causing damage or mortality and finally exit from the host [42]. All these mechanisms are aided by the expression of virulence factors at the different stages of infection. Several biochemical characteristics of the Vibrio isolates along with the analysis of their virulence factors and surface characteristics were used to determine the relationship between these factors and the pathogenic potential of the species. An essential step in the successful invasion of host requires adherence to host surfaces by flagella or pili [43,44] followed by production of extracellular polysaccharides eventually leading to biofilm formation [45]. Lytic enzymes produced by Vibrios enable them to procure nutrients present in the host and disseminate to different parts of host tissue. Among them, hemolysins, lipases, and gelatinases are well documented [46,47]. The ability of V.alginolyticus and V.parahaemolyticus strains mentioned in this study to produce multiple exo enzymes, as well as hemolysins make them prime contaminants in seafood. Experimental studies reveal the genes involved in chemotaxis also influence auto-aggregation and hydrophobicity of pathogenic bacteria. An interrelationship between these factors and biofilm is critical to expanding the knowledge regarding the development of biofilms in static and flowing aquatic environments [48].
Caenorhabditis elegans, a genetically traceable multicellular organism is an attractive host to address fundamental questions regarding pathogenicity of various Gram negative human pathogens. Our study looked into the pathogenic potential of isolated Vibrios using wild type C.elegans N2 strain. Vibrio alginolyticus infection often results in colonization of bacteria in worm gut within a short span of 8 hours [49]. The exposure of the nematodes for 24 hrs to three V. alginolyticus strains specified in our study also documented abnormalities and mortality due to the infection over a 10-day period. Virulent V.parahaemolyticus was reportedly associated with pharyngeal damage and distention leading to death of the nematode in 48 hours [50]. The worms fed on V.parahaemolyticus strain BTSV4 exhibited the lowest number of flings probably due to the same impact of the infection around the pharyngeal region.
Conclusion
The findings of the study support the conclusion that the presence of antibiotic resistant biofilm formers will compromise the safety and acceptability of seafood thereby co-equally endangering aquatic and human life. Furthermore, good aquaculture practices with special attention to food safety, environmental protection, and the well-being of farmed organisms must be implemented to safeguard food security and sanitation in future. An extensive elucidation of the molecular mechanisms involved in the infection or pathogenesis of Vibrios is mandatory to illustrate the notion of widespread contamination of antibiotics in shrimp aquaculture and remold new ways to combat this calamity.
Acknowledgement
The authors acknowledge Cochin University of Science and Technology, Kerala, India for supporting the work with necessary facilities.
References
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annual Reviews in Microbiology. 2002; 56: 187- 209.
- Flemming HC, Wingender J. The biofilm matrix. Nature reviews microbiology. 2010; 8: 623-633.
- Phillips CA. Bacterial biofilms in food processing environments: a review of recent developments in chemical and biological control. International Journal of Food Science & Technology. 2016; 51: 1731-1743.
- Holmström K, Gräslund S, Wahlström A, Poungshompoo S, Bengtsson BE, Kautsky N. Antibiotic use in shrimp farming and implications for environmental impacts and human health. International journal of food science & technology. 2003; 38: 255-266.
- Tang HJ, Chang MC, Ko WC, Huang KY, Lee CL, Chuang YC. In vitro and in vivo activities of newer fluoroquinolones against Vibrio vulnificus. Antimicrobial agents and chemotherapy. 2002; 46: 3580-3584.
- Akinbowale OL, Peng H, Barton MD. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. Journal of applied microbiology. 2007; 103: 2016-2025.
- Swapna KM, Rajesh R, Lakshmanan PT. Incidence of antibiotic residues in farmed shrimps from the southern states of India. 2012.
- Stalin N, Srinivasan P. Characterization of Vibrio parahaemolyticus and its specific phage from shrimp pond in Palk Strait, South East coast of India. Biologicals. 2016; 44: 526-533.
- Chang CI, Lee CF, Wu CC, Cheng TC, Tsai JM, Lin KJ. A selective and differential medium for Vibrio alginolyticus. Journal of fish diseases. 2011; 34: 227-234.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990; 215: 403-410.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987; 4: 406-425.
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular biology and evolution. 2007; 24: 1596-1599.
- Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. Journal of clinical pathology. 1989; 42: 872-874.
- Wolfe AJ, Millikan DS, Campbell JM, Visick KL. Vibrio fischeri s54 controls motility, biofilm formation, luminescence, and colonization. Appl Environ Microbiol. 2004; 70: 2520-2524.
- Laxmi M, Sarita G Bhat. Diversity Characterization of Biofilm Forming Microorganisms in Food sampled from Local Markets in Kochi, Kerala, India. International Journal of Recent Scientific Research. 2014: 5: 1070-1075.
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Burrett FF, Melton DM, et al. Adherence of coagulase negative staphylococcito plastic tissue culture plates: A Quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1988; 22: 996-1006.
- Stepanovic S, Vukovic D, Dakic I, Savic B, Švabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of microbiological methods. 2000; 40: 175-179.
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966; 45: 493-496.
- Murray B, Jorgensen L, Pfaller. Manual of clinical microbiology, 9thedition. American Society for Microbiology. 2007.
- Swift S, Lynch MJ, Fish L, Kirke DF, Tomás JM, Stewart GS, et al. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infection and immunity. 1999; 67: 5192-5199.
- Kos BV, Šuškovic J, Vukovic S, Šimpraga M, Frece J, Matošic S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. Journal of applied microbiology. 2003; 94: 981-987.
- Namdari HA, Bottone EJ. Correlation of the suicide phenomenon in Aeromonas species with virulence and enteropathogenicity. Journal of clinical microbiology. 1988; 26: 2615-2619.
- Janda JM, Oshiro LS, Abbott SL, Duffey PS. Virulence markers of mesophilic aeromonads: association of the autoagglutination phenomenon with mouse pathogenicity and the presence of a peripheral cell-associated layer. Infection and immunity. 1987; 55: 3070-3077.
- Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS microbiology letters. 1980; 9: 29-33.
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77: 71- 94.
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Current Biology. 2000; 10: 1539-1542.
- Sivamaruthi BS, Ganguli A, Kumar M, Bhaviya S, Pandian SK, Balamurugan K. Caenorhabditis elegans as a model for studying Cronobacter sakazakii ATCC BAA-894 pathogenesis. Journal of basic microbiology. 2011; 51: 540- 549.
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature reviews microbiology. 2004; 2: 95-108.
- Vert M, Doi Y, Hellwich KH, Hess M, Hodge P, Kubisa P, et al. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure and Applied Chemistry. 2012; 84: 377-410.
- Xie ZY, Hu CQ, Chen C, Zhang LP, Ren CH. Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Letters in applied microbiology. 2005; 41: 202-207.
- Vaseeharan B, Ramasamy P, Murugan T, Chen JC. In vitro susceptibility of antibiotics against Vibrio spp. and Aeromonas spp. isolated from Penaeus monodon hatcheries and ponds. International Journal of Antimicrobial Agents. 2005; 26: 285-291.
- Cai SH, Lu YS, Wu ZH, Jian JC, Wang B, Huang YC. Loop-mediated isothermal amplification method for rapid detection of Vibrio alginolyticus, the causative agent of vibriosis in mariculture fish. Letters in applied microbiology. 2010; 50: 480-485.
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in microbiology. 2009; 17: 109-118.
- Luo G, Huang L, Su Y, Qin Y, Xu X, Zhao L, et al. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerging Microbes & Infections. 2016; 5: e85.
- Marsden AE, Grudzinski K, Ondrey JM, DeLoney-Marino CR, Visick KL. Impact of salt and nutrient content on biofilm formation by Vibrio fischeri. PloS one. 2017; 12: e0169521.
- Han N, Mizan MF, Jahid IK, Ha SD. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food control. 2016; 70: 161-166.
- Ito A, Taniuchi A, May T, Kawata K, Okabe S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl Environ Microbiol. 2009; 75: 4093- 4100.
- Sengupta T, Sasmal D, Abraham TJ. Antibiotic susceptibility of luminous bacteria from shrimp farm environs of West Bengal. 2003.
- Sudha S, Divya PS, Francis B, Hatha AA. Prevalence and distribution of Vibrio parahaemolyticus in finfish from Cochin (south India). Vet Ital. 2012; 48: 281.
- Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression ofvps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. Journal of bacteriology. 2001; 183: 1716-1726.
- Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. Journal of bacteriology. 2005; 187: 752-757.
- Donnenberg MS. Pathogenic strategies of enteric bacteria. Nature. 2000; 406: 768-774.
- Chen Q, Yan Q, Wang K, Zhuang Z, Wang X. Portal of entry for pathogenic Vibrio alginolyticus into large yellow croaker Pseudosciaena crocea, and characteristics of bacterial adhesion to mucus. Diseases of Aquatic Organisms. 2008; 80: 181-188.
- Luo G, Huang L, Su Y, Qin Y, Xu X, Zhao L, et al. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerging Microbes & Infections. 2016; 5: e85.
- Muralidharan J, Jayachandran S. Physicochemical analyses of the exopolysaccharides produced by a marine biofouling bacterium, Vibrio alginolyticus. Process biochemistry. 2003; 38: 841-847.
- Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infection and immunity. 2010; 78: 1772-1780.
- Drake SL, DePaola A, Jaykus LA. An overview of Vibrio vulnificus and Vibrio parahaemolyticus. Comprehensive Reviews in Food Science and Food Safety. 2007; 6: 120-144.
- Min KR, Rickard AH. Coaggregation by the freshwater bacterium Sphingomonas natatoria alters dual-species biofilm formation. Appl Environ Microbiol. 2009; 75: 3987-3997.
- Durai S, Pandian SK, Balamurugan K. Establishment of a Caenorhabditis elegans infection model for Vibrio alginolyticus. Journal of basic microbiology. 2011; 51: 243-252.
- Durai S, Karutha Pandian S, Balamurugan K. Changes in Caenorhabditis elegans exposed to Vibrio parahaemolyticus. J Microbiol Biotechnol. 2011; 21: 1026-1035.