
Perspective
Austin J Microbiol. 2016; 2(1): 1013.
Chloramphenicol Peptides in the Ribosomal Tunnel
Vlachogiannis IA, Plessa E and Dinos GP*
Department of Biochemistry, University of Patras, Greece
*Corresponding author: Dinos GP, Department of Biochemistry, School of Medicine, University of Patras, 26500 Patras, Greece
Received: December 01, 2016; Accepted: December 01, 2016; Published: December 07, 2016
Keywords
Chloramphenicol-derivatives; Peptidyl-tRNA analogs; Nascent peptidyl-tRNA mimics; Ribosomal tunnel; Antibiotics
Perspective
Antibiotics kill bacteria by inhibiting essential enzymes involved in cellular metabolism, including also protein biosynthesis. However, extensive use and sometimes misuse of antibiotics have led to microbial resistance, which is constantly developing against all antibiotic classes, raising serious public health concerns and the urgency for the development of new antibacterial therapeutics [1]. Different strategies have been used to circumvent target-specific resistance, including finding new antibiotic targets and designing compounds with higher affinity to known targets [2]. A new scaffold for designing such new antibiotics, are peptide antibiotics.
Antimicrobial Peptides (AMPs) are a diverse group of molecules that play an important role in the innate immune response of plants and animals [3]. Having a minimum length of about thirty aminoacyl residues, AMPs often feature a net positive charge relay, due to a high lysine, arginine, and/or histidine content. This in turn, provides them with an amphiphilic character that enables them to associate with the phospholipid bilayer of bacteria, while staying clear from the eukaryotic cell membrane. Many AMPs form transmembrane pores that disorder the bacterial bilayer, causing rapidly lysis and cell death, an activity of particular concern when it comes to developing AMP-based therapeutics, given that high concentrations of peptide could finally result in unwanted cytotoxic effects on mammalian cells [4]. Other classes of peptides inhibit microbial growth by targeting intracellular processes, rather than damaging the bacterial membrane. Among these antimicrobials, Proline-Rich Antimicrobial Peptides (PrAMPs) have attracted high consideration in recent years as a possible way to manipulate the rapid increase in pathogen resistance [5,6]. Contrary to the pore-forming AMPs, PrAMPs are inserted into the bacterial cytoplasm by specific transporters [7], a transport mechanism absent in mammalian cells, and cross-react only unimportantly with intracellular eukaryotic components. As a result, they are generally accepted to be non-toxic [8], making them ideal scaffolds in developing novel antibacterial alternatives compared to classical antibiotics. Moreover, it is proven that certain PrAMPs, cross the blood-brain barrier, emphasizing their potential to be used in the future, as tissue-specific drug delivery systems.
For many years, it was thought that insect-derived PrAMPs exert their inhibitory effects by targeting the bacterial chaperone DnaK, a known heat shock protein [9]. However, recent papers have challenged this view, suggesting that ribosomal inhibition is key to defending against bacterial infection [10,11]. Soon after that, Xray crystal structure studies were published, revealing in depth the atomic details of the interactions between the bacterial 70S ribosome from T. thermophilus, and many PrAMPs like oncocin, bactenecin, metalnikowin I and pyrrhocoricin [11,12,13]. Oncocin is produced by the milkweed bug (Oncopeltus fasciatus) and is representative member of the previous entire family of PrAMPs [14]. The crystal structures as well as additional biochemical data revealed that PrAMPs interacts with the large subunit of the bacterial ribosome, occupying most of the functional ribosomal area starting from the aminoacyl-tRNA binding site up to the exit tunnel (Figure 1). More precisely, they block the binding of the incoming aminoacyl-tRNA, trapping the ribosome in an inactive initiation complex [11,12,13]. The binding site they occupied on the large ribosomal subunit is additionally overlapped with the well-known binding sites of many clinically important antibiotics, such as macrolides, pleuromutilins, chloramphenicols, and lincosamides [13,15].
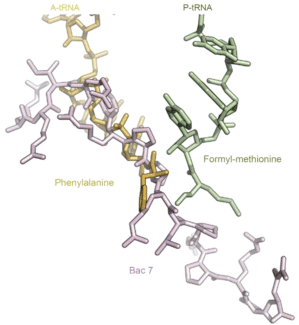
Figure 1: Crystal structure of antimicrobial peptide Bac 7 in the ribosomal exit
tunnel. Bac 7 (pink) binding site covers partially both the A-site and the P-site
areas and traps A-site aminoacyl-tRNAs in an inactive state. The molecular
structure was made using files PDB ID IVY4 [23] (for the tRNAs) and PDB ID
5F8K [11] (for Bac 7) and the Pymol software.
Experimental Procedure
Our research is oriented towards using chloramphenicol (CAM) as a trojan horse occupying the A-site in the Peptidyl Transferase Center (PTC) of 50S ribosomal subunit, with its dichloro-acetyl tail replaced by different peptidyl-residues, appropriate to insert in and interact with the ribosomal exit tunnel, as was previously resolved, using the same scaffold with shorter peptides [16]. These peptidyl derivatives of CAM were found to be tightly bound on the ribosome, occupying the A-site of the PTC, where chloramphenicol moiety is overlapped with the known antibiotic binding site, while the peptidylresidues were directed towards the exit tunnel. We appropriately modify the peptidyl-chain in both length and sequence, so that it could be inserted in and interact with residues of the exit tunnel. Taking into account that occupation of the exit tunnel resembles the macrolide antibiotic function, it is expected that peptidyl derivatives of CAM will act dually, not only as PTC inhibitors due to the CAM moiety, but additionally as antibiotics occupying the entrance of the tunnel thus occluding the route travel of nascent peptides from the PTC toward the exterior surface of the ribosome [17].
The proposed function of the ribosomal exit tunnel in the literature, had initially little to do with each specific nascent peptide sequence, serving exclusively as an exit corridor of peptide chains out of the ribosome. Now, it is well documented that specific aminoacid sequences of the nascent peptide chains interact with components of the exit tunnel, influencing many aspects of translation [18,19]. We use the established sequence of proline-rich peptides, especially the motif of Onc112 whose middle part (PYLPRP) binds to the PTC, an extremely conserved region of the ribosome and absolutely necessary for its function. Antibiotics bound to this region interact with nucleotide U2504 of 23S rRNA, that discriminates between bacterial and eukaryotic cells [11,13,20]. By adopting this motif as a necessary component of our constructs, we hope to ensure their specificity against bacteria. Additionally, we plan to extend the peptidyl-residue by changing the rest sequence of Onc112 looking not only for a more specific interaction, but for a more universal interaction with residues of the exit tunnel.
Many peptidyl derivatives of CAM have already been synthesized in our lab using solid-phase approaches [21], and some of them are proved to be interesting compounds and potential antimicrobial scaffolds.
Sooner or later, bacteria develop resistance to every antibiotic. When a selective advantage is raised, this resistance is readily transferred to other bacteria. Because of their effect over a wide variety of bacteria, broad-spectrum antibiotics provide a genetic selection for this transfer of resistance genes [1]. In addition, broad spectrum antibiotics disrupt the host microbiome and can result in chronic secondary problems [22]. Concerns about antibiotic side effects and resistance have been widely discussed in the scientific community and have recently received international attention as a serious global health problem. Therefore, the development of antibiotics with low toxicity and efficacy against resistant strains is a one-way road. Our procedure, offers a new scaffold for developing new antibiotics, that could combine low toxicity against human cells, higher potency and efficacy against resistant bacterial strains. While this is a very promising procedure for the development of novel antibiotics, it is not clear whether resistance caused by mutations that arise against currently used antibiotics will also confer cross-resistance against peptidyl derivatives of CAM. Nevertheless, the structural and biochemical knowledge that will be achieved through these studies could provide a solid background for the design of improved, peptidyl or peptidomimetic, antibacterial compounds [4].
Acknowledgement
We thank Dr. Stefan Arenz for helping us in figure production.
References
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014; 12: 35-48.
- Arenz S, Wilson DN. Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development. Mol Cell. 2016; 61: 3-14.
- Wang G, Mishra B, Lau K, Lushnikova T, Golla R, Wang X. Antimicrobial peptides in 2014. Pharmaceuticals (Basel). 2015; 8: 123-150.
- Wilson DN, Guichard G, Innis AC. Antimicrobial peptides target ribosomes. Oncotarget. 2015; 6: 16826-16827.
- Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. Apidaecins: antibacterial peptides from honeybees. EMBO J. 1989; 8: 2387-2391.
- Scocchi M, Tossi A, Gennaro R. Proline-rich antimicrobial peptides: converging to a nonlytic mechanism of action. Cell Mol. Life Sci. 2011; 68: 2317-2330.
- Runti G, Lopez Ruiz Mdel C, Stoilova T, Hussain R, Jennions M, Choudhury HG, et al. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7(1-35). J. Bacteriol. 2013; 195: 5343-5351.
- Hansen A, Schäfer I, Knappe D, Seibel P, Hoffmann R. Intracellular toxicity of proline-rich antimicrobial peptides shuttled into mammalian cells by the cell-penetrating peptide penetratin. Antimicrob Agents Chemother. 2012; 56: 5194-5201.
- Otvos L Jr, O I, Rogers ME, Consolvo PJ, Condie BA, Lovas S, et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000; 39: 14150-14159.
- Krizsan A, Volke D, Weinert S, Sträter N, Knappe D, Hoffmann R. Insect- Derived Proline-Rich Antimicrobial Peptides Kill Bacteria by Inhibiting Bacterial Protein Translation at the 70S Ribosome. Angew Chem Int Ed Engl. 2014; 53: 12236-12239.
- Seefeldt AC, Graf M, Pérébaskine N, Nguyen F, Arenz S, Mardirossian M, et al. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016; 44: 2429-2438.
- Seefeldt AC, Nguyen F, Antunes S, Pérébaskine N, Graf M, Arenz S, et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat Struct Mol Biol. 2015; 22: 470- 475.
- Roy RN, Lomakin IB, Gagnon MG, Steitz TA. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat Struct Mol Biol. 2015; 22: 466-469.
- Li W, Tailhades J, O’Brien-Simpson NM, Separovic F, Otvos L Jr, Hossain MA, et al. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino acids. 2014; 46: 2287-2294.
- Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann N Y Acad Sci. 2011; 1241: 1-16.
- Mamos P, Krokidis MG, Papadas A, Karahalios P, Starosta AL, Wilson DN, et al. On the use of the antibiotic chloramphenicol to target polypeptide chain mimics to the ribosomal exit tunnel. Biochimie. 2013; 95: 1765-1772.
- Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, et al. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014; 111: 15958-15963.
- Gupta P, Kannan K, Mankin AS, Vázquez-Laslop N. Regulation of gene expression by macrolide-induced ribosomal frameshifting. Mol Cell. 2013; 52: 629-642.
- Wilson DN, Arenz S, Beckmann R. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol. 2016; 37: 123- 133.
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014; 513: 517-522.
- Krokidis MG, Márquez V, Wilson DN, Kalpaxis DL, Dinos GP. Insights into the mode of action of novel fluoroketolides, potent inhibitors of bacterial protein synthesis. Antimicrob Agents Chemother. 2014; 58: 472-480.
- Blaser Martin. Missing Microbes: How the Overuse of Antibiotics Is Fueling Our Modern Plagues, Henry Holt and Co, New York, 2014.
- Polikanov YS, Steitz TA, Innis CA. Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. Nat Struct Mol Biol. 2014; 21: 787-793.