
Research Article
Austin Med Sci. 2022; 7(3): 1071.
Prognostic and Predictive Value of Aberrant P53 Status in Human Cancers: Systematic Review and Meta-Analysis
Luo X#, Wang HK#, Guan LL, Yang YY#, Meng XR and Wang F*
Department of Oncology, The First Affiliated Hospital of Zhengzhou University, China
#These authors contribute equally to this work and share first authorship
*Corresponding author: Feng Wang Department of Oncology, The First Affiliated Hospital of Zhengzhou University, No.50 Eastern Jianshe Road, Zhengzhou 450052, Henan Province, China
Received: November 14, 2022; Accepted: December 19, 2022; Published: December 25, 2022
Abstract
Background: Many studies have attempted to correlate p53 abnormality including p53 gene mutation and p53 protein over expression with prognosis or therapeutic response in adjuvant chemotherapy but have yielded controversial results. To investigate whether p53 aberrations have different impacts on survival and outcomes of adjuvant chemotherapy among cancer patients, we conducted a meta-analysis.
Methods: We performed comprehensive search before September 18, 2022. Hazard Ratio (HR) with 95% Confidence Interval (CI) was effective measure, and Stata 16.0 was applied for data analysis.
Results: A total of 14 articles were enrolled in this meta-analysis. p53 protein over expression detected by immunohistochemistry was a risk factor of 5-year Overall Survival (OS) among cancer patients after Radical Surgery (RS). Furthermore, p53 protein over expressed patients displayed inferior response to adjuvant chemotherapy with unfavorable 5-year Disease Free Survival (DFS) (HR = 1.61, 95% CI: 1.12 ~ 2.32, P = 0.011). p53 gene mutation was a negative indicator of OS in adjuvant chemotherapy (HR = 1.41, 95% CI: 1.19 ~ 1.69, P< 0.001). Furthermore, we performed subgroup analysis according to year of publication, the number of patients, detection method of abnormal p53 and tumor types, consistent result was observed.
Conclusion: p53 protein over expression appears to serve as a predictive and prognostic biomarker in adjuvant chemotherapy setting. p53 gene mutation is a potential predictor and could identify high-risk patients who obtain limited overall benefit from adjuvant chemotherapy and support clinical decision-making.
Keywords: p53; Adjuvant chemotherapy; Cancer; Prognosis; Meta-analysis
Abbreviations: OS: Overall Survival; DFS: Disease Free Survival; HR: Hazard Ratio; CI: Confidence Interval; RS: Radical Surgery; MDM2: Murine Double Minute 2; IHC: Immunohistochemical Staining; NSCLC: Non-Small Cell Lung Cancer; CRC: Colorectal Cancer; Wtp53: Wild-Type P53; Mutp53: Mutant P53; GOF: Gain-Of- Function; PCNA: Proliferating Cell Nuclear Antigen; EGFR: Epidermal Growth Factor Receptor; MDR1: Multiple Drug Resistance 1.
Introduction
Cancer, a worldwide devastating disease with high incidence, poses a great threat to human health and existence. According to data from the International Agency for Research on Cancer, there were 19.3 million new cases and 10 million cancer deaths worldwide in 2020 [1]. In addition, the incidence of cancer is expected to increase in coming years. Treatment regimens for cancers include conventional surgery, chemotherapy, radiotherapy, molecular targeted therapy, immunotherapy, traditional Chinese medicine and their combination [2-7]. Despite impressive progress in tumor therapy, local recurrence and distant metastasis account for the main causes of mortality [8]. Adjuvant chemotherapy, the adjunction of chemotherapy after RS, significantly reduces the risks of relapse and death by killing residual tumor cells. It is often recommended as the best therapeutic option for stage colorectal cancer (TanyN1-2M0) [9,10], hormone- unresponsive breast cancer [11] stage B-A non-small cell lung cancer (NSCLC) [12] after RS. Though the preponderance of adjuvant chemotherapy has been clearly established, the benefit of adjuvant chemotherapy varies widely among patients with the same tumor type and a substantial proportion of patients acquire resistance and relapse. Therefore, it is urgent and critical to develop biomarkers that could accurately assess high-risk patients who mostly respond to adjuvant chemotherapy and determine optimum therapeutic regimen.
The critical p53 tumor suppressor gene, located on human chromosome 17p13.1, encodes a 375 amino acid nuclear phosphoprotein [13]. p53 protein plays a variety of roles in the prevention and suppression of tumor initiation and progression through cell cycle arrest, cellular apoptosis, cellular senescence, DNA repair, inhibition of angiogenesis, and so on [14,15]. Mutations in the p53 gene are frequently documented in more than half of all human cancers [16-18] and majority of mutations are missense mutations in DNA binding domains, a region that is necessary for wild-type p53 (wtp53) to bind to its target genes and perform its function as a transcription factor [19,20]. While the critical role of wtp53 in tumor suppression has been firmly established, a growing body of evidence has demonstrated that mutant p53 (mutp53) proteins not only lose the tumorsuppressive function of wtp53, but also confers novel activities to promote tumorigenesis independently of wtp53, referred to as gain-of-function (GOF) [21]. One of the characteristics of mutp53 proteins is that mutp53 proteins are prone to more stable and accumulate in tumors which are primarily mediated by dysfunctional murine double minute 2 (MDM2). MDM2 is a key negative regulator for wtp53. MDM2 binds to and degrades wtp53 through ubiquitination. Meanwhile, MDM2 itself is a p53-regulated gene. In this way, MDM2 forms a negative feedback loop with wtp53 p53 to tightly regulate wtp53 p53 protein levels and functions [22-25]. In light of this characteristic of the mutp53 protein, positive Immunohistochemical staining (IHC) of p53 in tumor tissues has been widely used as a surrogate for detection of mutp53. However, p53 protein overexpression is not always equal to p53 gene mutation in several tumors. A plethora of studies reported that p53 protein overexpression occur frequently in a broad range of cancers [26,27]. To date, dozens of studies have focused on the relationship between p53 abnormalities and adjuvant chemosensitivity, but the conclusions are still inconsistent to a great extent. For instance, Popat et al [28] reported that p53 overexpression increases the sensitivity of adjuvant chemotherapy among patients with Colorectal Cancer (CRC), whereas Williams et al [29] suggested that p53 overexpression does not affect the chemosensitivity of patients with CRC in adjuvant setting.
Meta-analysis is generally considered a powerful statistical tool to overcome the limitations of different sample sizes from individual studies to generate the best overall estimation. Therefore, the study aims to elaborate precisely the relationship between the p53 alteration and prognosis, as well as effectiveness of adjuvant chemotherapy among cancer patients. Hoping our study will provide essential and valuable information for clinicians in clinical practice.
Materials and Methods
Search Strategy
The meta-analysis was carried out in accordance with the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement and methods. A comprehensive search was conducted to identify relevant studies by two independent reviewers (XL, HKW). We retrieved all relevant literature published in English from four online databases consisting of PubMed, Embase, Web of Science and Cochrane Library until September 18, 2022. HR and 95% CI for OS and DFS was extracted. Following keywords were used for the search: (“TP53 Genes” OR “p53 Genes” OR “p53 expression” OR “p53 protein” OR “p53 accumulation” OR “p53 status” OR “p53 overexpression”) AND (“Adjuvant Chemotherapy” OR “Drug Therapy, Adjuvant” OR “Adjuvant Drug Therapy” OR “Postoperative chemotherapy”) AND (“carcinoma” OR “tumor” OR “neoplasia” OR “neoplasm”, “cancer” OR “malignancy” OR “malignant neoplasm”).
Inclusion and Exclusion Criteria
In order to achieve our research objectives, we determined the following inclusion and exclusion criteria. The inclusion criteria were as following: (1) prospective or historical cohort studies; (2) cancer patients receiving adjuvant chemotherapy after RS or only RS; (3) p53 alteration either detected as overexpression in the protein level or as mutation by the DNA level; (4) correlation between p53 and survival indexes, such as 5-year OS and DFS; (5) providing HR with 95% CI or data to calculate. Articles were excluded if one of the following criteria was fulfilled: (1) articles with insufficient sample size (n <30); (2) animal studies, case report, reviews, comments, and editorials; (3) duplicate data or analysis; (4) insufficient data for calculating HR and 95% CI; (5) literature not published in English.
Data Extraction and Quality Evaluation
The following data were extracted from all included studies: (1) name of the first author; (2) the year of publication; (3) the country; (4) tumor types; (5) sample size of study; (6) agents of adjuvant chemotherapy; (7) detection method for p53 gene mutation or p53 protein expression; (8) primary outcome measures including 5-year OS and DFS; (9) HR of OS and DFS. Two investigators (XL, HKW) conducted data extraction independently. An external referee (LLG) was involved in case of disagreements which could not be resolved by both investigators. The data was summarized in extraction table (Table 1) and analyzed manually. The quality of all eligible studies was assessed by two investigators (LLG, YYY) according to the Newcastle-Ottawa Scale (NOS) independently. The NOS consists of three parts: selection (0-4points), comparability (0-2 points), and outcome assessment (0-3 points). NOS scores of ≥ 6 were regarded as highquality studies. The NOS scores of all included articles were ≥ 6, and the scoring details are presented in (Table 2).
Study
Year
Country
Tumor types
Adjuvant chemotherapy
Detection of p53 alteration
Outcome measures
Adjuvant/ total patients
NOS scores
Elsaleh
1999
Australia
rectal cancer
5-FU /levamisole
PCR - SSCP
OS
35/122
6
Elsaleh
2000
Australia
colon cancer
5-FU /levamisole
PCR - SSCP
OS
133/388
6
Soong
2000
Australia
colorectal cancer
5-FU /levamisole
PCR - SSCP
OS
154/894
7
Powell
2001
Australia
breast cancer
CMF - based
PCR - SSCP
OS
172/586
8
Anderson
2005
Sweden
breast cancer
CMF - based
PCR - SSCP
OS
125/376
8
Tsao
2007
Canada
NSCLC
Cisplatin / vinorelbine
gene sequencing / IHC
OS
242/482
8
Ma
2014
France
NSCLC
Cisplatin-based
gene sequencing
OS / DFS
264/524
8
Kandioler
2015
Austria
colon cancer
5-FU - based
gene sequencing
OS
389/389
7
Li
2018
China
colorectal cancer
5-FU - based
gene sequencing
OS
177/315
7
Malamou
2006
Greece
breast cancer
CMF - based
IHC
OS / DFS
392/392
8
Popat
2006
Britain
colorectal cancer
5-FU - based
IHC
OS
766/967
8
Dumontet
2010
France
breast cancer
TAC or FAC
IHC
OS / DFS
1334/1334
7
Graziano
2010
America
NSCLC
carboplatin or paclitaxel
IHC
OS / DFS
124/250
8
Williams
2019
Australia
colorectal cancer
5-FU - based
IHC
DFS
189/264
7
PCR: Polymerase Chain Reaction; SSCP: Single Strand Conformation Polymorphism; IHC: Immunohistochemistry; OS: Overall Survival; DFS: Disease Free Survival; CMF: Cyclophosphamide / Methotrexate / 5-Fluorouracil; 5-FU: 5-Fluorouracil; NSCLC: Non-Small Cell Lung Cancer; FAC / TAC: Fluorouracil Or Docetaxel / Doxorubicin / Cyclophosphamide.
Table 1: Characteristics of the included studies in meta-analysis.
Study
Selection
Comparability
Exposure
Scores
Representativeness of the exposed cohort
Selection of the noneexposed cohort
Ascertainment of exposure
Demonstration that outcome of interest was not present at start of study
Comparability of cohorts on the basis of the design or analysis
Assessment of outcome
Was follow-up long enough for outcomes to occur
Adequcy of follow up of cohorts
Elsaleh (1999)
*
*
*
*
**
-
-
-
6
Elsaleh (2000)
*
*
*
*
**
-
-
-
6
Soong (2000)
*
*
*
*
**
-
*
-
7
Powell (2001)
*
*
*
*
**
*
*
-
8
Andersson (2005)
*
*
*
*
**
-
*
*
8
Tsao (2007)
*
*
*
*
**
-
*
*
8
Ma (2004)
*
*
*
*
**
-
*
*
8
Kanioler (2015)
*
*
*
*
**
-
*
-
7
Li (2018)
*
*
*
*
**
-
*
-
7
Popat (2006)
*
*
*
*
**
*
*
-
8
Malamou (2006)
*
*
*
*
**
-
*
*
8
Dumontet (2010)
*
*
*
*
**
-
*
-
7
Grazino (2010)
*
*
*
*
**
-
*
*
8
Williams (2019)
*
*
*
*
**
-
-
*
7
A maximum of one ‘*’ for each item within the ‘Selection’ and ‘Exposure’ categories, and maximum of ‘**’ for the ‘Comparability’. If the score is 6 or greater, the literature is considered to be of high quality.
Table 2: Characteristics of the included studies in meta-analysis.
Statistical Analysis
Data analysis was calculated by using Stata version 16.0 software (Stata Corporation, College Station, TX, USA). We conducted a pooled analysis of HR for 5-year OS and DFS. The I² statistic and Q test were used to examine between-study heterogeneity. When there was substantial heterogeneity (I²> 50%, P < 0.05) in the study, the random-effects models were adopted. Otherwise, we applied the fixed-effects models as the pooling method. If necessary, the sensitivity analysis and subgroup analysis were performed. All reported P-values were two-sided, and the P-values less than 0.05 were considered significant.
Results
Literature Search and Study Characteristics
Of the four databases, there were 723 records after duplicates were removed. Subsequently, 633 records were omitted by screening titles and abstracts. For the remaining 90 records, 76 articles were excluded by reading full-text articles. At last, the remaining 14 articles involving 6083 individuals were included in this meta-analysis. A flow diagram with details of the study selection process was presented in (Figure 1). Among the 14 studies, 7 studies focused on colorectal cancer; 4 studies focused on breast cancer; 3 studies focused on NSCLC. HR and 95% CI was extracted directly from 14 studies. IHC was applied to detect the expression of p53 in all included studies. p53 mutation was detected by polymerase chain reaction-single strand conformation polymorphism or gene sequencing. The main characteristics of the eligible studies were summarized in (Table 1).
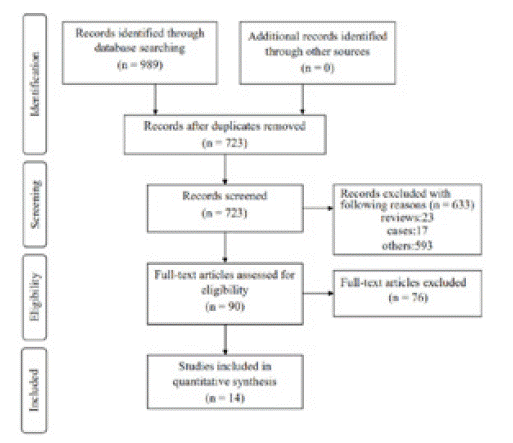
Figure 1: The flowchart of study selection process.
The Prognostic Value of Abnormal P53 Status among Patients after RS
In order to explore the prognostic significance of abnormal p53 status in cancers, a total of 7 articles were finally included in this pooled analysis. HR with 95% CI for 5-year OS was aggregated by using fixed-effects models or random-effects models. Results of pooled analysis from 5 studies illustrated that there was no significant improvement in 5-year OS (HR = 1.06, 95% CI: 0.83 ~ 1.37, P = 0.633) (Figure 2) between patients containing mutp53 and those without, revealing mutp53 is not a potential prognostic factor in patients with cancer. Considering that p53 protein overexpression is not always consistent with p53 gene mutation, we further investigated the interaction between p53 protein overexpression and prognosis in cancers. Two studies reported the relationship between p53 protein overexpression and OS among NSCLC patients receiving RS, respectively. Though it is impossible to pool these results, both studies indicated that p53 protein overexpression could predict worse 5-year OS among patients with NSCLC after RS (HR = 1.89, 95% CI: 1.07 ~ 3.34; HR = 2.30, 95% CI: 1.44 ~ 3.68). In brief, p53 protein overexpression, not mutp53, seems to serve as an unfavorable indicator in NSCLC patients.
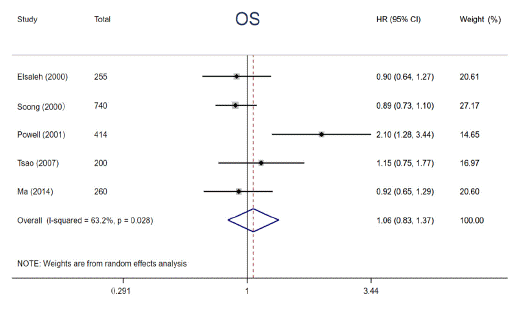
Figure 2:
The Predictive Significance of Mutp53 in Adjuvant Chemotherapy
Numerous studies have reported that p53 gene mutations have distinct impacts on effects of adjuvant chemotherapy, although the results are disputed. In the study, 8 eligible studies were enrolled to elucidate the impact of p53 mutation on 5-year OS among cancer patients treated with adjuvant chemotherapy. We performed a pooled analysis by using fixedeffects models due to limited heterogeneity (I² = 0, P = 0.960). The pooled results demonstrated that patients with p53 gene mutation derived no significant survival benefit from adjuvant chemotherapy for 5-year OS (HR = 1.41, 95% CI: 1.19 ~ 1.69, P< 0.001) (Figure 3), whereas those with wild type p53 did. Subgroup analysis stratified by year of publication, the number of patients, detective method of abnormal p53 and tumor types was conducted, the results didn’t alter across different subgroups (Figure 4). Notably, this phenomenon is more pronounced in patients with Colorectal Cancer (CRC) (HR = 1.48, 95% CI: 1.16 ~ 1.90, P = 0.002) compared with non-CRC (HR = 1.34, 95% CI: 1.04 ~ 1.73, P = 0.024). The above results supported that p53 mutation status is perceived as a negative predictive factor and interferes with the effects of adjuvant chemotherapy among cancer patients especially in CRC.
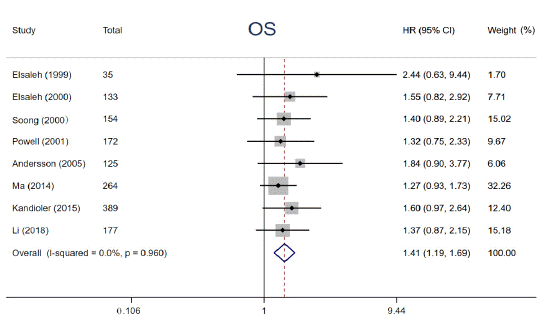
Figure 3:
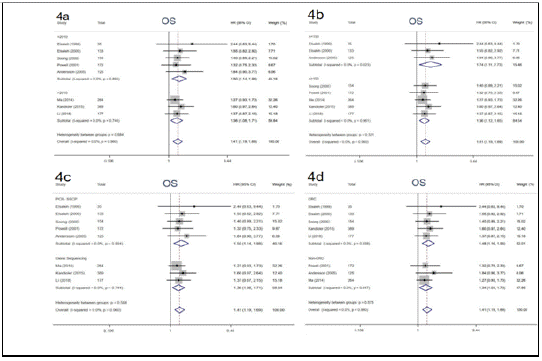
Figure 4:
The Predictive Significance of P53 Protein Overexpression in Adjuvant Chemotherapy
p53 protein overexpression, encoded by p53 gene, presents in a variety of tumors and exerts an undesirable influence on tumor progression and drug resistance. Prior studies have demonstrated that p53 protein accumulation in cancer has been found to be correlated with diverse impacts on adjuvant chemotherapy. To clarify the relationships, relevant data from 6 articles were extracted and analyzed. The data were calculated by using a random-effects models owing to its substantial heterogeneity (I² = 73.6%, P = 0.023). The patients harboring p53 protein overexpression appeared a distinctly decreased 5-year DFS compared to those without (HR = 1.61, 95% CI: 1.12 ~ 2.32, P = 0.011) (Figure 5). In addition, a trend of shorter 5-year OS among patients carrying p53 protein overexpression in adjuvant chemotherapy setting was also observed, however, the difference was not statistically significant (HR = 1.49, 95% CI: 0.90 ~ 2.46, P = 0.122) (Figure 6). p53 protein overexpression could be regarded as an essential determinant in adjuvant setting and is a potential biomarker for selecting cancer patients who are inclined to resist to adjuvant chemotherapy.
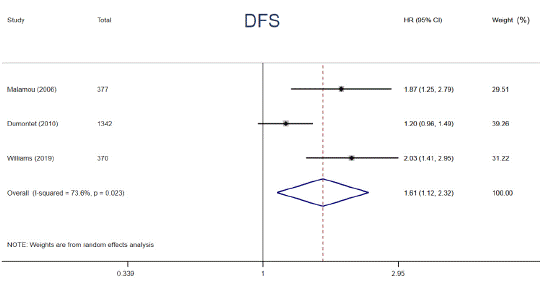
Figure 5:
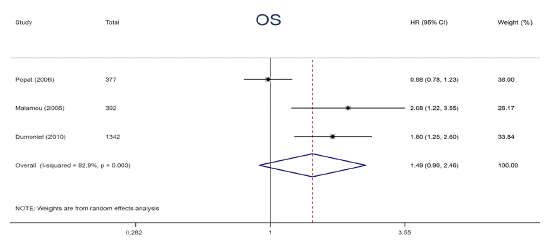
Figure 6:
Sensitivity Analysis
The sensitivity analysis was applied to assess whether OS was interfered by individual studies in cancer patients harbored mutp53 in adjuvant chemotherapy setting. Sensitivity analysis demonstrated that there was no remarkable influence on the merged results when any individual study was removed. At last, we discovered that the HR values of OS and the synthesized results were stable and reliable (Figure 7).
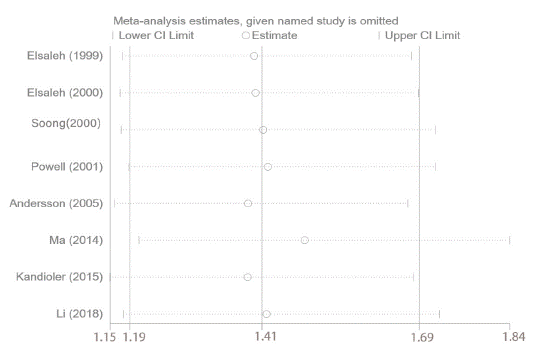
Figure 1:
Discussion
Cancer has a tremendous influence on patients, their families, and the society. With the development of cancer diagnostic and therapeutic technologies, patient survival time has been significantly extended in the last few decades. p53 gene is consistently one of the most frequently mutated genes detected in all types of cancer. Mutant p53 proteins are often stabilized and significantly overexpressed in many tumor tissues through multiple mechanisms including MDM2 short isoforms [30,31], BAG family proteins [32,33], chaperone proteins Hsp90 [34,35], etc. However, mutp53 proteins lack inherent stability in normal cells/tissues. The phenomenon indicates that cancer cells promote the stability and accumulation of mutp53 protein that is a critical premise of mutp53 GOF. In addition, multiple studies have reported that p53 gene mutations and elevated p53 expression are frequently associated with more aggressive cancer, poorer response to adjuvant chemotherapy, and prognosis in several different types of human cancer [36,37].
The majority of patients harbor p53 mutations are accompanied by high levels of p53 protein. p53 protein overexpression, however, is not consistent with the presence of p53 mutations gene in many cancers such as in oral squamous cell carcinoma, lymphomas and infiltrative ductal breast carcinoma implicating the existence of other underlying mechanisms [38-40]. Thus, this is the first meta-analysis to investigate the association between p53 gene mutations and elevated p53 expression and the prognosis of cancer patients, as well as the benefit of adjuvant chemotherapy, respectively. Taken together, our combined results indicated that p53 protein overexpression, not p53 gene mutation, had significant prognostic effect and correlated with inferior 5-year OS rate among cancer patients received RS. A marginal predictive effect for adjuvant chemotherapy was observed for DFS. Furthermore, patients with p53 gene mutations trend to exhibit poor 5-year OS and are resistant to adjuvant chemotherapy. Due to high incidence of p53 mutations in CRC patients, we performed a subgroup analysis according to tumor types, the finding is consistent and more pronounced in patients with CRC.
In our study, we conclude that mutp53is not a prognostic indicator among cancer patients receiving RS. The definition of mutp53 is conservative and mutp53 is not further divided which might attenuate the significance of mutp53 for prognosis. For example, non-missense mutations of p53 (including nonsense, splice site and frame shift mutation) have little effect on wtp53 activity and the survival of cancer patients. Our results suggested patients with mutp53 trended toward limited survival benefit from adjuvant chemotherapy. To explain this association, it would be interesting to hypothesize that these p53 mutants may exert a GOF effect that interferes with response to adjuvant chemotherapy. So far, several underlying mechanisms of mutp53 GOFs have been identified and presented as following. (1) mutp53 in general could not recognize and bind to p53 DNAbinding elements and lose the transcriptional activity of wtp53 [41]; (2) mutp53 proteins could form a tetramer with wtp53, which may block the function of the remaining wtp53 in tumor suppression [42]. (3) mutp53proteins could bind to p63 and p73 [43,44], both two important members of p53 family, and inhibit their antitumor activities; (4) mutp53 could enhance tumor cell survival and its ability to resist adjuvant chemotherapy through multiple metabolic pathways such as aerobic glycolysis [45], oxidative phosphorylation [46], mevalonate pathway and sterol biosynthesis [47]; (5) mutp53 could regulate the transcription of some genes [48-51] such as Proliferating Cell Nuclear Antigen (PCNA), Epidermal Growth Factor Receptor (EGFR) and Multiple Drug Resistance 1 (MDR1) via different mechanisms, which promote tumor cell proliferation, migration, invasion and chemoresistance. High levels of p53 expression corelates with poor prognosis and efficacy of adjuvant chemotherapy. Accumulation of mutp53 protein promotes mutp53 GOF and malignant progression of cancer which may account for reduced survival after RS and worse response to adjuvant chemotherapy.
In summary, it is possible that p53 alteration is recommended as an essential and reliable biomarker of adjuvant chemotherapy to individualize treatment, and to achieve maximal efficacy among cancer patients, especially in patients with CRC. More recently, p53 has emerged as an attractive cancer therapeutic target. Given that mutp53 proteins often accumulate to high levels and exert a GOF effect to promote malignant progression in human cancer, targeting mutp53 has become an attractive therapeutic strategy for cancer harbor mutp53 proteins. The primary strategies for targeting mutp53 are restoring the wtp53 p53 activity and depleting mutp53 protein in cancers. Combined application of small molecule drugs that restore wtp53 activity or deplete mutp53 proteins could enhance the sensitivity of postoperative adjuvant chemotherapy, improve therapeutic efficacy and prolong survival time among cancer patients with mutp53. Despite our best efforts to conduct a comprehensive analysis, our meta-analysis has some limitations. First, we analyzed the prognostic and predictive value of p53 abnormalities across a variety of tumors, which may not be applicable to patients with a certain tumor type. Second, the scale of the included studies was limited. Third, the cutoff values that define p53 overexpression by IHC are variable among these studies. Therefore, further analysis with rigid criteria and large-scale prospective studies are warranted to define the relationship between abnormal p53 and prognosis, as well as effects of adjuvant chemotherapy.
Conclusion
In brief, our analysis revealed that elevated p53 expression and p53 gene mutation seem to be useful molecular biomarkers for prognostication, risk stratification, and selection of cancer patients especially in patients with CRC most likely to benefit from adjuvant chemotherapy. Further studies with large populations are desperately needed to confirm our findings.
Authors’ Contributions
XL and HKW: Literature search, Data collection and analysis, and Writing manuscript; LLG: Validation, Investigation and Resources. YYY: Formal analysis and Data Curation. XRM: Methodology, Writing a reviewing and Editing. FW: Supervision and Funding acquisition. All authors read and approve the final manuscript prior to submission.
Conflicts of Interest and Source of Funding
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. This work was supported by the National Natural Science Funds of China (No. 81672442) and the Scientific and Technological Projects in Henan Province (No. 182102310379).
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021; 71: 209-249.
- Salibasic M, Pusina S, Bicakcic E, Pasic A, Gavric I, Kulovic E, et al. Colorectal Cancer Surgical Treatment, our Experience. Med Arch. 2019; 73: 412-414.
- Kim JH. Chemotherapy for colorectal cancer in the elderly. World J Gastroenterol. 2015; 21: 5158-5166.
- Chua B, Jackson JE, Lin C, Veness MJ. Radiotherapy for early nonmelanoma skin cancer. Oral Oncol. 2019; 98: 96-101.
- Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 2018; 834: 188-196.
- Zhang X, Qiu H, Li C, Cai P, Qi F. The positive role of traditional Chinese medicine as an adjunctive therapy for cancer. Biosci Trends. 2021; 15: 283-298.
- Wang S, Long S, Wu W. Application of Traditional Chinese Medicines as Personalized Therapy in Human Cancers. Am J Chin Med. 2018; 46: 953-970.
- Cho WK, Roh JL, Cho KJ, Choi SH, Nam SY, Kim SY. Lymph node ratio predictive of recurrence, distant metastasis, and survival in submandibular gland carcinoma patients. J Cancer Res Clin Oncol. 2019; 145: 1055-1062.
- Teufel A, Gerken M, Fürst A, Ebert M, Hohenthanner I, Klinkhammer- Schalke M. Benefit of adjuvant chemotherapy in high-risk colon cancer: A 17-year population-based analysis of 6131 patients with Union for International Cancer Control stage II T4N0M0 colon cancer. Eur J Cancer. 2020; 137: 148-160.
- Saltz LB. Adjuvant therapy for colon cancer. Surg Oncol Clin N Am. 2010; 19: 819-827.
- Lluch A, Barrios CH, Torrecillas L, Ruiz-Borrego M, Bines J, Segalla J, et al. Phase III Trial of Adjuvant Capecitabine After Standard Neo-/Adjuvant Chemotherapy in Patients with Early Triple-Negative Breast Cancer (GEICAM/2003-11_CIBOMA/2004-01). J Clin Oncol. 2020; 38: 203-213.
- Kubota K, Kunitoh H, Seto T, Shimada N, Tsuboi M, Ohhira T, et al. Randomized phase II trial of adjuvant chemotherapy with docetaxel plus cisplatin versus paclitaxel plus carboplatin in patients with completely resected non-small cell lung cancer: TORG 0503. Lung Cancer. 2020; 141: 32-36.
- Chang F, Syrjänen S, Kurvinen K, Syrjänen K. The p53 tumor suppressor gene as a common cellular target in human carcinogenesis. Am J Gastroenterol. 1993; 88: 174-186.
- Panatta E, Zampieri C, Melino G and Amelio I. Understanding p53 tumour suppressor network. Biol Direct. 2021; 16: 14.
- Machado-Silva A, Perrier S, Bourdon JC. p53 family members in cancer diagnosis and treatment. Semin Cancer Biol. 2010; 20: 57- 62.
- Kandioler D, Schoppmann SF, Zwrtek R, Kappel S, Wolf B, Mittlbock M, et al. The biomarker TP53 divides patients with neoadjuvantly treated esophageal cancer into 2 subgroups with markedly different outcomes. A p53 Research Group study. J Thorac Cardiovasc Surg. 2014; 148: 2280-2286.
- Kaur RP, Vasudeva K, Kumar R and Munshi A. Role of p53 Gene in Breast Cancer: Focus on Mutation Spectrum and Therapeutic Strategies. Curr Pharm Des. 2018; 24: 3566-3575.
- Li H, Zhang J, Tong JHM, Chan AWH, Yu J, Kang W, et al. Targeting the Oncogenic p53 Mutants in Colorectal Cancer and Other Solid Tumors. Int J Mol Sci. 2019; 20: 5999.
- Muller PA and Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014; 25: 304-317.
- Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers?. Cell Death Differ. 2018; 25: 154-160.
- Schulz-Heddergott R and Moll UM. Gain-of-Function (GOF) Mutant p53 as Actionable Therapeutic Target. Cancers (Basel). 2018; 10: 188.
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006; 21:307-315.
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005; 24: 2899-2908.
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013; 13: 83-96.
- Henningsen KM, Manzini V, Magerhans A, Gerber S, Dobbelstein M. MDM2-Driven Ubiquitination Rapidly Removes p53 from Its Cognate Promoters. Biomolecules. 2021; 12: 22.
- Auvinen A, Isola J, Visakorpi T, Koivula T, Virtanen S, Hakama M. Overexpression of p53 and long-term survival in colon carcinoma. Br J Cancer. 1994; 70: 293-296.
- Kim KW, Kim N, Choi Y, Kim WS, Yoon H, Shin CM, et al. Different effects of p53 protein overexpression on the survival of gastric cancer patients according to Lauren histologic classification: a retrospective study. Gastric Cancer. 2021; 24: 844-857.
- Popat S, Chen Z, Zhao D, Pan H, Hearle N, Chandler I, et al. A prospective, blinded analysis of thymidylate synthase and p53 expression as prognostic markers in the adjuvant treatment of colorectal cancer. Ann Oncol. 2006; 17: 1810-1817.
- Williams DS, Mouradov D, Browne C, Palmieri M, Elliott MJ, Nightingale R, et al. Overexpression of TP53 protein is associated with the lack of adjuvant chemotherapy benefit in patients with stage III colorectal cancer. Mod Pathol. 2020; 33: 483-495.
- Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002; 2: 9-15.
- Harris LC. MDM2 splice variants and their therapeutic implications. Curr Cancer Drug Targets. 2005; 5: 21-26.
- Yue X, Zhao Y, Huang G, Li J, Zhu J, Feng Z, et al. A novel mutant p53 binding partner BAG5 stabilizes mutant p53 and promotes mutant p53 GOFs in tumorigenesis. Cell Discov. 2016; 2: 16039.
- Yue X, Zhao Y, Liu J, Zhang C, Yu H, Wang J, et al. BAG2 promotes tumorigenesis through enhancing mutant p53 protein levels and function. Elife. 2015; 4: e08401.
- Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011; 18: 1904-1913.
- Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1998; 18: 1517-1524.
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010; 2: a001008.
- Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012; 33: 2007-2017.
- Ara N, Atique M, Ahmed S and Ali Bukhari SG. Frequency of p53 gene mutation and protein expression in oral squamous cell carcinoma. J Coll Physicians Surg Pak. 2014; 24: 749-753.
- Martinez-Delgado B, Robledo M, Arranz E, Infantes F, Echezarreta G, Marcos B, et al. Correlation between mutations in p53 gene and protein expression in human lymphomas. Am J Hematol. 1997; 55: 1-8.
- Pezeshki AM, Farjadian S, Talei A, Vasei M, Gharesi-Fard B, Doroudchi M, et al. p53 gene alteration and protein expression in Iranian women with infiltrative ductal breast carcinoma. Cancer Lett. 2001; 169: 69-75.
- Schulz-Heddergott R, Moll UM. Gain-of-Function (GOF) Mutant p53 as Actionable Therapeutic Target. Cancers (Basel). 2018; 10: 188.
- Milner J, Medcalf EA, Cook AC. Tumor suppressor p53: analysis of wild-type and mutant p53 complexes. Mol Cell Biol. 1991; 11: 12-19.
- Stindt MH, Muller PA, Ludwig RL, Kehrloesser S, Dotsch V and Vousden KH. Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene. 2015; 34: 4300-4310.
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002; 416: 560-564.
- Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013; 4: 2935.
- Basu S, Gnanapradeepan K, Barnoud T, Kung CP, Tavecchio M, Scott J, et al. Mutant p53 controls tumor metabolism and metastasis by regulating PGC-1alpha. Genes Dev. 2018; 32: 230-243.
- Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez- Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012; 148: 244-258.
- Polotskaia A, Xiao G, Reynoso K, Martin C, Qiu WG, Hendrickson RC, et al. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci U S A. 2015; 112: E1220-1229.
- de Rozieres S, Maya R, Oren M and Lozano G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene. 2000; 19: 1691-1697.
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009; 139: 1327-1341.
- Chin KV, Ueda K, Pastan I and Gottesman MM. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science. 1992; 255: 459-462.