
Research Article
Austin Med Sci. 2019; 4(1): 1033.
Knockdown of PARP1 Inhibits β-Lapachone-Mediated Reductions in AGS Cell Proliferation and Migration
Wang C1*, Li C4*, Jin W2, Lin Z3, Quan X2,5# and Shen X2#
1Department of Interventional Medicine, Jilin Cancer Hospital, China
2Department of Oncology, Yanbian University Affiliated Hospital, China
3Cancer Research Center and Department of Pathology, Yanbian University Medical College, China
4Department of Gastrology, Yanbian University Affiliated Hospital, China
5Department of Postdoctoral Programme, Yanbian University Affiliated Hospital, Yanji, Jilin, China *Chu Wang and Chenghao Li contributed equally to this work
*Corresponding author: Xionghu Shen and Xianglan Quan, Department of Oncology, Yanbian University Affiliated Hospital, Yanji, Jilin, China
Received: July 01, 2019; Accepted: August 14, 2019; Published: August 21, 2019
Abstract
Background: β-lapachone selectively induces apoptotic cell death in a variety of human cancer cells in vitro and in vivo. However, the mechanism by which β-lapachone promotes cell death in gastric cancer is not fully understood. The present study aimed to investigate the role and molecular mechanism of PARP1 in the β-lapachone-mediated killing of the human gastric carcinoma cell line AGS.
Methods: AGS cells were transfected with non-specific or target-specific small interfering RNAs (siRNAs) using riboFECTTM CP. After transfection, each of the 4 groups was treated with 4 μmol/l β-lapachone, and western blotting was used to detect changes in the protein expression of the PARP1 cleavage product. Variance analysis was used to compare the results of the 4 treatment groups, and the most stably transfected cell lines were selected. The best siRNA was selected for subsequent cell transfections, and a negative control group with NC-siRNA was also established.
Results: AGS cell proliferation and colony-forming ability were detected by the MTT and colony formation assays, respectively, whereas AGS cell migration was detected by the scratch assay. Knockdown of PARP1 via siRNA transfection did not change the cells’ morphology. The expression of cleaved PARP1 protein was significantly reduced in cells with PARP1 knockdown. Treatment with β-lapachone markedly abolished the proliferative, colony-forming and migratory abilities of the control AGS cells but did not affect AGS cells with PARP1 knockdown.
Conclusion: Our novel findings suggest that PARP1 has an important role in the β-lapachone-mediated killing of gastric cancer cells.
Keywords: Gastric cancer; PARP1; β-lapachone
Background
In China, gastric cancer has the second highest incidence among malignant tumors. Gastric cancer has a high rate of metastasis and high mortality with reported metastasis and recurrence rates of 33% after radical gastrectomy [1]. A recent study shows that treatment with radiotherapy and chemotherapy can result in drug resistance due to the survival of solid tumors in a hypoxic environment; this ultimately leads to treatment failure [2]. Recently, some drugs, including β-lapachone, streptonigrin and deoxynyboquinones (DNQs), have been developed to specifically target tumor cells that can survive in hypoxic conditions [3]. β-lapachone is primarily extracted from the bark of red sandalwood trees from Central and South America; additionally, it possesses antibacterial, antifungal, and antitumor properties and has extensive biological activity [5]. Nevertheless, β-lapachone can not only inhibit the activity of cancer cells but also induce tumor cell apoptosis [4]. Moreover, β-lapachone can enhance NQO1-mediated toxicity in cells and exert anti-cancer activities [5].
The PARP family contains 18 subtypes, with PARP1 as the predominantly expressed subtype. PARP1 was first extracted from the nuclei of liver cells in 1963 by a French group led by Mandel. The important role of PARP1 in DNA damage monitoring, DNA repair, transcriptional regulation, chromatin modification and so on has attracted the attention of many scholars. In addition, PARP1 is highly expressed in breast cancer, liver cancer, gastric cancer, lymphoma, colon cancer, ovarian cancer and other malignant tumor cells and plays a key role in tumor progression [6-8]; furthermore, PARP1 is also involved in cell death pathways [9,10]. Previous studies reported that β-lapachone induces apoptosis in gastric cancer cells via the classical caspase pathway, and one study observed that PARP1 was overactivated and cleaved during the study [11]. It remains unclear, however, whether PARP1 is involved in the β-lapachone-mediated killing of gastric cancer cells and the mechanism by which this occurs. In this study, we observed that knocking down PARP1 with siRNA transfection did not change the cells’ morphology. The expression of cleaved PARP1 protein was significantly reduced in AGS cells with PARP1 knockdown. Furthermore, treatment with β-lapachone markedly abolished the proliferative, colony-forming and migratory abilities of AGS cells but did not affect cells with PARP1 knockdown.
Materials and Methods
Cell cultures
AGS cells were supplied by the Cancer Research Center of Yanbian University and cultured in a humidified atmosphere (37°C) containing 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) with RPMI 1640 (Gibco, Thermo Fisher Scientific, China) supplemented with 10% FBS (Gibco), Standard Serum Supplementation, and 4500 mg/L glucose. The growth of AGS cells was observed under a microscope until the AGS cells were adherent, after which the cells were treated promptly.
siRNA transfection
pAGS cells were transfected with non-targeting or targetspecific small interfering RNA (siRNA) using riboFECTTM CP (Guangzhou Rui Bo Biological Technology). siRNAs targeting PARP1 were designed and synthesized by Guangzhou Ruibo Biotechnology. Three unique PARP1 siRNA sequences were constructed as follows: 1, 5’-GGAGGAAGGTATCAACAAA-3’; 2, 5’-GGGCAAGCACAGTGTCAAA-3’; and 3, 5’-GGAACAAGGATGAAGTGAA-3’. The NC-siRNA sequence was 5’-TTCTCCGAACGTGTCACGT-3’. AGS cells were suspended in complete medium, seeded in 6-well plates at 8 x 105 cells / well, and divided into the following 4 groups: PARP1-siRNA 1 transfection group, PARP1-siRNA 2 transfection group, PARP1-siRNA 3 transfection group and NC-siRNA (negative control) group. When the AGS cells were 30 to 50% confluent, the siRNA mixtures were diluted and prepared with riboFECTTM CP, which were then added to the cell culture medium and mixed. The plates were incubated at 37°C in an atmosphere containing 5% CO2, and the infection rate was assessed under a microscope 48 h after transfection. Then, 4 μmol/ L β-lapachone was added to each group for subsequent western blotting.Western blot analysis
Cells from the 4 treatment groups were lysed and processed to produce protein extracts that were subjected to SDS-PAGE, membrane transfer, antibody hybridization and ECL chemiluminescence detection to detect the expression of the PARP1 cleavage product. A primary antibody for cleaved PARP1 was used, and quantitative data were analyzed using NIH ImageJ software.
MTT assay
For the MTT assay, siRNA-transfected cells were incubated with 3-(4,5-dimethylhioazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT (50 μg/well) for 4 hrs and then treated with dimethylsulfoxide (100 μl/well). The absorbance (A570-A630) of each well was measured by using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony formation assay
Cells in the logarithmic growth phase and that had been treated with β-lapachone were digested with trypsin. After the cells were dissociated and centrifuged, they were resuspended in cell culture medium. The cell suspension was serially diluted and seeded in culture dishes at a density of 5000 cells/dish. The culture dishes were gently shaken to evenly distribute the cells and were placed in an incubator for 2 weeks. Cell growth was observed periodically until colonies were visible to the naked eye, at which point the supernatant was discarded, and 2 ml of anhydrous formaldehyde per well as added for 15 min to fix the cells. Next, GIMSA staining was performed on the cells for 10 ~ 30 min, after which the residual dye was washed off, and the plate was dried at room temperature. The culture dish was inverted on transparent stage, and the colonies were counted either at a low magnification microscopy or with the naked eye to calculate the colony formation rate.
Scratch assay
The bottom of a 6-well plate was marked with a pen and ruler to draw a straight line with the average width of 0.5 ~ 1 cm. Cells from the PARP1-siRNA and NC-siRNA transfection groups were seeded into the 6-well plate. When the cells reached 90% confluence, scratches were carefully made through the monolayer with a 10-μl pipette tip to avoid tilt. After the cells were washed three times with PBS and then administered serum-free medium, they were treated with 4 μmol/L β-lapachone and placed in an incubator (5% CO2, 37°C). Photographs of the marks were taken at 0, 12, 24, and 48 h after scratching, and the distance between the scratches was measured under a microscope.
Statistical analyses
The data were analyzed by SPSS 18.0 statistical software. The data were expressed as x±s. Using analysis of variance, the experimental data between the two groups were compared by t-test. All experiments were repeated 3 times, and p‹0.05 indicated statistically significant differences.
Results
Transfection efficiency of AGS gastric cancer cell lines
AGS cells were transfected with siRNA for 48 hours. The morphology of the cells (round, adherent growth) as observed under a microscope showed no significant changes compared with the control group, indicating that silencing PARP1 has no significant effect on cell morphology and growth (data not shown).
Western blot of the PARP1 cleavage protein
To understand the effect of silencing PARP1 after on the killing effects of β-lapachone, we treated AGS cells that were transfected with siRNA with 4 μmol/L β-lapachone and assessed PARP1 expression via western blotting. Compared with the control group, the three PARP1 siRNA groups showed significant reductions in the expression of PARP1 cleavage protein (Figure 1, p‹0.05). Cells transfected with PARP1-siRNA1 exhibited an 89% decrease (Figure 1, p‹0.01); PARP1-siRNA2, a 73% decrease (Figure 1, p‹0.05), and PARP1- siRNA3, an 83% decrease (Figure 1, p‹0.05). These data indicate that PARP1-siRNA 1 was the most effective sequence to silence PARP1 gene expression and was thus used for all subsequent experiment
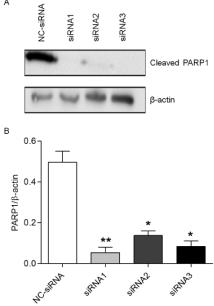
Figure 1: AGS cells transfected with control or PARP1-specific siRNAs
were treated with 4 μmol/L β-lapachone, and western blotting was used to
compare the changes in the expression of the PARP1 cleavage product.
Comparisons were conducted using the t-test. The expression of the PARP1
cleavage product was significantly decreased in the PARP1-siRNA1 (89%,
p‹0.01), PARP1-siRNA2 (73%, p‹0.05), and PARP1-siRNA3 (83%, p‹0.05)
transfection groups compared to the levels in the NC-siRNA group. Data are
means from 3 independent experiments, in triplicate. *p‹0.05, **p‹0.01.
Effects of PARP1-siRNA transfection on AGS cell proliferation, colony formation and migration
To examine the impact of PARP1 knockdown on β-lapachoneinduced cell death, we assessed the proliferative, colony-forming and migratory abilities of AGS cells with PARP1 knockdown via the MTT, colony formation and scratch assays, respectively. The results showed that the inhibition of the growth rate of NC-siRNA cells gradually increased with increasing concentrations of β-lapachone (Figure 2) after 24 h; when the concentration was 4 μmol/L, the growth rate of AGS cells was inhibited by approximately 50%. However, the inhibitory effects of β-lapachone in the PARP1-siRNA-transfected cells were significantly decreased (Figure 2, p ‹0.05).
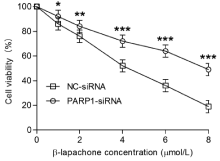
Figure 2: The MTT assay was used to compare the proliferation of AGS cells
in the PARP-siRNA and NC-siRNA groups. The results show that the inhibition
of the growth rate in NC-siRNA cells gradually increased with increasing
concentrations of β-lapachone (1, 2, 4, 6, 8 μmol/L) after a 24-h treatment.
When the drug concentration was 4 μmol/L, the growth inhibition rate of AGS
cells was approximately 50%. However, the inhibitory effects of β-lapachone
on the growth of PARP1-siRNA-transfected cells were significantly decreased
(p‹0.05). Data are presented as the means from 3 independent experiments
conducted in triplicate. *p‹0.05, **p‹0.01, ***p‹0.001.
The colony formation assay showed that among AGS cells treated with 4 μmol/L β-lapachone for 2 weeks, cells in the NC-siRNA group had fewer colonies, whereas those in the PARP1-siRNA group had a significant increase in the number of colonies formed, which corresponded to a 1.5-fold increase in the colony formation rate (Figure 3, p ‹0.05). These results suggesting that β-lapachone can significantly inhibit the proliferative and colony-forming ability of AGS cells via PARP1 and that excessive activation of PARP1 plays a key role in the pathogenesis of β-lapachone to kill gastric cancer cells.
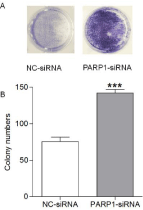
Figure 3: The effect of β-lapachone on the colony-forming ability of AGS
cells subjected to PARP1-siRNA and NC-siRNA transfection was detected
by the colony formation assay. When AGS cells were treated with 4 μmol/L
β-lapachone for 2 weeks, cells in the NC-siRNA group had fewer colonies,
whereas those in the PARP1-siRNA group had significant more colonies, and
the colony formation rate was 1.5 times higher than that in the NC-siRNA
group (p‹0.05). Data are presented as the means from 3 independent
experiments conducted in triplicate. ***p‹0.001.

Figure 4: AGS cells transfected with PARP1-siRNA and NC-siRNA were
administered 4 μmol/L β-lapachone and subjected to the scratch assay. At
0, 12, 24 and 48 hours after scratching, the width of the scratches in both
groups was observed and measured. The healing distance of the PARP1-
siRNA transfection group was significantly shorter than that of the NC-siRNA
group and was time-dependent (p‹0.05).
Discussion
Gastric cancer is the most common malignancy of the digestive system. Four subtypes lead in the incidence of malignant tumors, and two subtypes account for the two highest mortality rates [12]. Although treatment options for gastric cancer (i.e., surgery, radiotherapy and chemotherapy) and other related research has made profound progress, the prognosis of patients with gastric cancer has not significantly improved.
The lack of selectivity of chemotherapy drugs for most cancer cells remains a major limiting factor. In recent years, many studies have focused on the observation of elevated NQO1 expression in most solid tumors, such as gastric and breast cancer; thus, drug research and development has been centered around NQO1 [13]. β-lapachone, a quinone chemotherapeutic agent, is an effective inhibitor of NQO1 and not only inhibits the biological activity of tumor cells but also the production of reactive oxygen species (ROS) to induce apoptosis of gastric cancer and other solid malignancies [14].
Extensive NQO1-mediated single-strand breaks (SSBs) cause poly(ADP-ribose) polymerase-1 (PARP1) hyper activation, which inhibits essential base and SSB repair functions. PARP1 hyper activation also causes dramatic reduction in the NAD+/ATP pool due to ADP ribosylation, resulting in extensive energy depletion and cell death. Therefore, β-lapachone is an attractive experimental drug.
PARP1 is a highly abundant ribozyme that plays a key role in repairing DNA damage, maintaining genome integrity, and regulating transcriptional translation levels [15]. In recent years, studies focusing on the association between PARP1 and tumorigenesis, how regulating PARP 1 can improve the effectiveness of tumor treatments and other aspects of related research have made rapid progress. X. Huang and others noted that the induced lethality of β-lapachone is mediated by NQO1-mediated oxidative reduction. When NQO1+ and NQO1- clones of the non-small cell lung cancer cell line H596 and the breast cancer cell line MDA-MB-231 were treated with DNQ (a quinone chemotherapeutic agent), DNQ could completely kill the NQO1+ cells but was ineffective against NQO1- cells. Similarly, in the NQO1+ human prostate cancer cell line PC-3, knocking down NQO1 using siRNA elicited the same effects as cell treated with dicoumarol (dicoumarol Inhibits NQO1), can inhibit the lethality of DNQ. In this study, PARP1 expression was knocked down with a specific siRNA, and the extent of NAD+ and ATP depletion was significantly decreased. Inhibition of PARP1 activation resulted in a significant reduction in programmed necrosis of tumor cells that were treated with DNQ for 24 h (P ‹0.05). These data suggest that β-lapachone acts through NQO1 and that gastric cancer cells die in response to PARP1 overexpression.
In this study, we silenced PARP1 in AGS cells and investigated the effects of β-lapachone effect on proliferation, colony formation and migration. We also explored the role of PARP1 in the destructive effects of β-lapachone on gastric cancer cells and the mechanism involved. The levels of PARP1 cleavage products in PARP1 siRNAtransfected cells was significantly decreased. The initial results suggest that the transfection of PARP1-siRNA was successful. We also observed that β-lapachone caused the overexpression of PARP1 and its cleavage products in normal gastric cancer cells, which is consistent with previous studies. The results of the MTT assay showed that AGS cell proliferation was significantly decreased in cells transfected with of PARP1-siRNA. The survival rate of AGS cells decreased with increasing concentrations of β-lapachone and decreased significantly at a dose of 4.0 μmol/L. The results of colony formation assay showed that the colony-forming ability of cells transfected with PARP1- siRNA was significantly higher than that in the control cells after treatment with 4.0 μmol/L β-lapachone (p‹0.05). The scratch assay results showed that the scaffold healing distance in the PARP1- siRNA transfection group was significantly shorter than that in the control group and was time-dependent (p‹0.05), Taken together with the abovementioned experimental results, our data indicate that β-lapachone will lead to excessive intracellular PARP1 activation and cleavage and eventual cell death in normal gastric cancer cells; furthermore, if PARP1 expression is knocked down, there is a reduction in PARP1 cleavage products—this mitigates the effects of β-lapachone on gastric cancer cell lethality.
One study noted [16] that a series of responses in the classical caspase pathway regulate the entire process of apoptosis, and different caspases vary in their regulation and mechanisms. The results of this experiment show that the expression of PARP1 cleavage protein in PARP1-siRNA-transfected cells was significantly lower than that in the NC-siRNA control group (p‹0.05). The proliferative, colonyforming and migratory abilities of NC-siRNA cells were significantly affected by the β-lapachone treatment, whereas the PARP1-siRNA exhibited completely opposing responses. This observation indicates that overactivation of PARP1 and excessive production of its cleavage products are involved in β-lapachone-mediated apoptosis of gastric cancer cells. When we incorporate the previous research results and the results of this study, we can conclude that the β-lapachonemediated killing of gastric cancer cells relies on excessive PARP1 activation; activated PARP1 is cleaved, and the cleavage product induces apoptosis by participating in the caspase pathway.
One prerequisite for apoptosis is that the cells contain certain levels of ATP and NAD+. When the DNA structure is widely and severely damaged, caspases can also cleave PARP1 to mediate apoptosis. PARP1 alone cannot induce apoptosis, but PARP1 in the presence of caspases is important for apoptosis [17]. Caspase cleavage of PARP1 prevents full-length PARP1 from initiating its DNA repair function and using adenosine diphosphate in the process of glycosylation, which consumes a large amount of ATP; blocking PARP1 activity ensures the smooth completion of the process of apoptosis. PARP1 is activated immediately after the DNA damage occurs. Although there are high levels of PARP1 during initial DNA breakage, there is only a small amount of PAR during the final stages of apoptosis. The cleaved PARP1 products play a key role in apoptosis.
Our findings herein demonstrate that PARP1 protein expression in AGS cells was significantly increased after β-lapachone treatment, and the proliferative, colony-forming and migratory abilities were significantly decreased concomitant with an increased cell death rate. When PARP1 was knocked down by siRNA, the expression of PARP1 cleavage products was significantly lower than that in cells transfected with NC-siRNA. Meanwhile, β-lapachone inhibited the proliferation, colony formation and migration of AGS cells, and the cell death rate was significantly increased. When the expression of PARP1 (and its cleavage products) was knocked down, the lethality of β-lapachone was also significantly reduced; indicating that over activated PARP1 plays a key role in the β-lapachone-mediated killing of gastric cancer cells. Previous studies have clarified [18] that β-lapachone kills tumor cells through the NQO1-mediated inhibition of redox reactions and oxygen consumption, which results in the accumulation of Reactive Oxygen Species (ROS). Increased levels of ROS cause extensive DNA damage. When the DNA is slightly damaged, PARP1 uses NAD+ as the substrate to catalyze the glycosylation reaction of adenosine diphosphate to form PAR, which can further guide DNA damage repair.
Conclusion
In conclusion, β-lapachone can significantly inhibit the proliferation, colony formation and migration of AGS gastric cancer cells by activating PARP1 to induce cell death. When we used siRNA knock down PARP1 in AGS cells, the protein expression of PARP1 was significantly reduced. Significant inhibition of PARP1 activation will significantly reduce the sensitivity of AGS cells to β-lapachone. PARP1 plays a key role in the β-lapachone-mediated killing effects on gastric cancer cells, and elucidating this mechanism has extensive research potential regarding the development of future antitumor drugs.
Acknowledgments
The authors thank Dr. Xiong Hu. Shen for supplying β-lapachone and the Imaging and Chemistry and Cancer Shared Resources, SCC.
Funding
state natural sciences fund (81560478).
References
- Bensaad K, Harris A L. Hypoxia and metabolism in cancer. Advances in Experimental Medicine & Biology. 2014; 772: 1.
- Ptok H, Gastinger I, Meyer F, et al. Hospital volume effects in surgical treatment of gastric cancer. Der Chirurg. 2016: 1-9.
- Parkinson EI, Hergenrother PJ. Deoxynyboquinones as NQO1-Activated Cancer Therapeutics. Accounts of Chemical Research. 2015; 48: 2715-2723.
- Wu Y, Wang X, Chang S, Lu W, Liu M, Pang X. β-lapachone Induces NQO1- and Oxidative Stress-Dependent Hsp90 Cleavage and Inhibits Tumor Growth and Angiogenesis. Journal of Pharmacology & Experimental Therapeutics. 2016; 357: 466.
- Kee JY, Han YH, Kim DS, Mun JG, Park SH, So HS, et al. β-lapachone suppresses the lung metastasis of melanoma via the MAPK signaling pathway. Plos One. 2017; 12.
- Cui X, Li L, Yan G, Meng K, Lin Z, Nan Y, et al. High expression of NQO1 is associated with poor prognosis in serous ovarian carcinoma. BMC Cancer. 2015; 15: 244.
- Liang S, Esswein SR, Ochi T, Wu Q, Ascher B, Sibanda L, et al. Achieving selectivity in space and time with DNA double-strand-break response and repair: molecular stages and scaffolds come with strings attached. Structural Chemistry. 2017; 28: 161-171.
- Ran C. Free energy calculation provides insight into the action mechanism of selective PARP-1 inhibitor. Journal of Molecular Modeling. 2016; 22: 1-9.
- Hazra J, Mukherjee P, Ali A, Poddar S, Pal M. Engagement of Components of DNA-Break Repair Complex and NF-KB in Hsp70A1A Transcription Upregulation by Heat Shock. Plos One. 2017; 12: e0168165.
- Narne P, Pandey V, Simhadri PK, Phanithi PB. Poly (ADP-ribose) Polymerase-1 hyperactivation in neurodegenerative diseases: the death knell tolls for neurons. Seminars in Cell & Developmental Biology. 2017; 63: 154.
- Katona BW, Rustgi AK. Gastric Cancer Genomics: Advances and Future Directions: Cellular & Molecular Gastroenterology & Hepatology. 2017; 3: 211-217.
- Meade JC, Moore ZR, Stuart SH, Cholka A. Abstract B39: NQO1 as a therapeutic target for atypical teratoid/ rhabdoid tumor. Cancer Research. 2016; 76: B39-B39.
- Mohindra NA, Patel JD. Towards manageable toxicities from targeted lung cancer treatment. Lung Cancer Management. 2017; 4: 279-287.
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016; 353: 45-50.
- Huang X, Dong Y, Bey EA, Kilgore JA, Bair JS, Li LS, et al. An NQO1 substrate with potent antitumor activity that selectively kills by PARP1- induced programmed necrosis. Cancer Research. 2012; 72: 3038-3047.
- Kerr LE, Mcgregor AL, Amet LE, Asada T, Spratt C, Allsopp TE, et al. Mice overexpressing human caspase 3 appear phenotypically normal but exhibit increased apoptosis and larger lesion volumes in response to transient focal cerebral ischemia. Cell Death & Differentiation. 2004; 11: 1102.
- Yu S W, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, et al. Mediation of Poly(ADP-Ribose) Polymerase-1-Dependent Cell Death by Apoptosis-Inducing Factor. Science. 2002; 297: 259-263.
- Daugherty MD, Young JM, Kerns JA, Mallik HS. Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts. Plos Genetics. 2014; 10: e1004403.s.