
Research Article
Ann Hematol Oncol. 2025; 12(1): 1473.
Effect Of SNHG6/Mir-579-3p/TFRC Regulatory Network on Haemoglobin H-CS Disease
Wang X1#, Li Y1#, Chen G1, Kashif A1, Mo L1, Li J1, Hou W2, Zhu C1*
1Genetics and Precision Medicine Laboratory, Affiliated Hospital of Guilin Medical University, Guilin-541001, China
2NHC Key Laboratory of Thalassemia Medicine, Guangxi Key Laboratory of Thalassemia Research, Life Sciences Institute, Guangxi Medical University, Nanning, Guangxi-530021, China
#Xinyu Wang and Yulan Li Contributed Equally to this Work
*Corresponding author: Chunjiang Zhu, Genetics and Precision Medicine Laboratory, Affiliated Hospital of Guilin Medical University, Guilin-541001, China Email: zhuchunjiang@glmc.edu.cn
Received: February 11, 2025; Accepted: March 06, 2025; Published: March 10, 2025;
Abstract
Background: Hemoglobin H-Constant Spring disease (HbH-CS disease) is the most common non-deletion HbH disease in the south of China, and it is an autosomal recessive blood system disease. The interaction of long non-coding RNAs (lncRNAs), which have been shown to interact with microRNAs (miRNA) could provide new insights into the epigenetic regulation of bead conversion and potentially develop new therapeutic modalities for thalassemia. The aim of this experiment was to investigate the effect of SNHG6/miR-579-3p/TFRC regulatory network on hemoglobin H-CS disease.
Methods: Bioinformatics analysis was used to predict the potential target genes miRNAs and miRNA-targeted mRNAs of SNHG6, and Real Time Quantitative Polymerase Chain Reaction (RT-qPCR) was used to analyse the differential expression of lncRNAs, miRNAs and mRNAs. Dual luciferase reporter gene assay was performed to identify the targeting relationship between SNHG6, miRNA, and mRNA. SNHG6-mediated oxidative stress and iron death effects were detected after loss-of-function and gain-of-function effects.
Results: SNHG6 is capable of interacting as a competitive endogenous RNA (ceRNA) with a variety of miRNAs, including miR-579-3p, and its competitive interaction with miR-579-3p regulates the expression level of TFRC. Silencing of SNHG6 was able to reduce the expression level of TFRC and decrease the level of cellular oxidative stress and iron death. Silencing miR-579-3p was able to restore SNHG6-mediated oxidative stress and iron death levels.
Conclusion: Our study confirms that SNHG6 activates TFRC expression through competitive interaction with miR-579-3p, which affects haemoglobin and iron death levels in haemoglobin H-CS disease. It provides new insights into the development of haemoglobin H-CS disease and may provide new targets for the treatment of patients with haemoglobin H-CS disease.
Keywords: Thalassemia; HbH-CS; SNHG6; miR-579-3p; TFRC
Introduction
Thalassaemia is one of the most common single-gene disorders and is an inherited haemolytic disorder with reduced production of haemoglobin pearl protein chains due to mutations or deletions in the pearl protein gene, with a- and Β-thalassaemia being the most common types [1]. Common in the southern region of China, Guangdong, Guangxi and Hainan, with the rate of carrier of the causative gene in Guangxi region being the highest in the country at 23.9 per cent, of which a-thalassaemia is 17.5 per cent and Β-thalassaemia 6.4 per cent, which is a major health problem in Guangxi Province [2,3].
Normally, human haemoglobin is a tetramer consisting of 2 pairs of bead protein chains, with the gene coding for a-bead protein located near the telomere on chromosome 16, including 1 embryonic and 2 fetal/adult genes along the chromosome [4]. Humans have four genes coding for alpha-pearl proteins, and when three of these genes are mutated or missing, alpha-globin chain synthesis is reduced, and excess beta-globin chains form a tetramer, known as hemoglobin H disease (HbH disease) [5]. HbH diseases can be divided into deletional and non-deletional types, with haemoglobin H-Constant Spring disease (HbH-CS disease) being the most common nondeletional type of alpha-thalassaemia [6]. HbH-CS disease is caused by a non-deletion-type mutation in the a2- globin chain gene that disrupts the stop codon and results in an unstable a- globin chain [7]. Compared with deletional types of HbH diseases, patients with HbH-CS disease develop growth defects in infancy, have more severe anemia, and haemolytic crises caused by infections, fever, etc., and often require regular blood transfusions [8].
Long non-coding RNA (lncRNA) is a non-coding RNA composed of more than 200 nucleotide units. lncRNAs play important roles in chromatin modification, transcriptional regulation, etc., in which they are important for the protection and stabilisation of mRNAs, and are able to competitively bind with specific miRNAs to organise the binding of miRNAs to their target mRNAs, thus relieving the inhibitory effect of miRNAs on gene synthesis [9,10]. It has been shown that lncRNAs are involved in the regulation of the terminal maturation stage of erythrocytes during their proliferation and differentiation [11]. With the intensive study of lncRNAs, several lncRNAs were found to be differentially expressed in thalassaemia, which may be involved in the proliferation and differentiation of the haematopoietic system and Hb production [12-14]. However, the role and mechanism of lncRNAs in HbH-CS disease are not clear. The aim of this study was to investigate the expression profile of lncRNAs in HbH-CS disease and whether lncRNAs play a role in biological processes such as haemoglobin synthesis and erythrocyte proliferation and differentiation in HbH-CS disease in the hope of providing new approaches and strategies for the treatment of HbH-CS disease.
Methods
Study Participants
In this experiment, peripheral blood samples were collected from four HbH-CS patients (genotype: --SEA /aCSa) and four normal (Hb = 110 g/L) healthy subjects. The exclusion criteria of the present study were as follows:1) concomitant with other thalassemia mutations; 2) serum ferritin (SF) = 12ug/ul; 3) received splenectomy; 4) history of blood transfusion within two months; 5) concomitant with infections, neoplasms, and autoimmune disorders. The study complied with the Declaration of Helsinki Principles and was approved by the Ethics Committee of the Affiliated Hospital of Guilin Medical University (NO.2023YJSLL-156). Participants were from the Affiliated Hospital of Guilin Medical University and provided written informed consent before the start of the experiment.
Cell Culture and Transfection
The cell lines used in this experiment were human chronic myeloid leukaemia K562 cells (MeilunBio, China) and human embryonic kidney 293T cell line. The K562 cell line stably expressing SNHG6 and miR-579-3p was established by lentiviral infection (Genechem, China). Cells were cultured in RPMI1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 ug/ml streptomycin in an incubator at 37°C, 5% CO2.
Separation of Nucleated Red Blood Cells
About 10 ml of peripheral blood sample was collected, and monocytes were isolated using density gradient centrifugation Monocyte Separation Medium (Solarbio, China). Reticulocytes and nucleated erythrocytes were separated from monocytes using the CD71+ (CD71 Antibody, anti-human, REAfinity™) selection method of the MACSTM separation system (Miltenyi Biotech, Auburn, CA, USA), and the purity of the sorting was determined by flow cytometry.
lncRNA Microarray Analysis
After isolation of nucleated erythrocytes from the collected 4V4 samples, they were analysed by Arraystar microarray sequencing, and differentially expressed lncRNAs between the two comparison groups were identified by fold change (FC) and statistical significance (P-value) thresholds (Kangcheng Biotechnology, China).
Bioinformatics Analysis
Arraystar microarray raw data were screened with P-value < 0.05 and FC > 1.5. The Starbase [15] was used to predict potential target genes of SNHG6 and miR-579-3p to construct ceRNA networks.
RNA Extraction and RT-qPCR
Total RNA was extracted using TRIzol Reagent (Takara, Japan), and then reverse transcribed to cDNA using PrimeScript RT Enzyme Mix I kit and miRNA fluorescence quantitative RT kit (Takara, Japan). RT-qPCR analysis was carried out using TB Green Premix Ex Taq II kit (Takara, Japan) for RT-qPCR analysis. Relative gene expression was calculated using the 2-ΔΔCt comparison method with Β-actin as the internal reference gene. Primer sequences are shown in Table 1.

Table 1: Primer sequences of related genes in RT-qPCR.
Dual-luciferase Reporter Gene Assay
According to the prediction of Starbase database, there are binding sites between SNHG6 and miR-579-3p, and between miR- 579-3p and TFRC. After constructing the seed sequences, they were inserted into wild-type (WT) and mutant (MUT) plasmids by doubleenzyme digestion, and the mimics and negative controls (NC) were co-transfected into cells with wild-type or mutant plasmids. The luciferase activity in the cell lysate was measured using a dual luciferase reporter kit (Hanbio Biotechnology, China) and a microplate reader.
Determination of Lipid Oxidation (MDA) Content
MDA content was determined using the Lipid Oxidation (MDA) Assay Kit (Beyotime, China). The standards were configured and standard curves were made according to the instructions. A total of 4×106 cells were taked and 0.4 ml of PBS was added, followed by 0.2 ml of MDA detection working solution, heated in boiling water bath for 30 min, cooled to room temperature, and centrifuged at 1000g/10min. The supernatant was taken and added into 96-well plate (200μl/well), and the absorbance value at 532 nm was determined. All the experiments were carried out independently for three times.
Determination of Reduced Glutathione (GSH) Content
The intracellular GSH content was determined using the Elabscience Reduced Glutathione (GSH) Colorimetric Assay Kit (Elabscience, China). Standards were configured and standard curves were made according to the instructions. One ×106 cells were taken and 300 μl of PBS was added, mixed well and centrifuged at 10,000 g/10 min at 4°C, then the supernatant was retained for measurement. A total of 0.1ml of supernatant and 0.1ml of Acid Reagent were mixed well, centrifuge at 4500g/10min and left to be measured.
One hundred μl of supernatant and 100 μl of Acid Reagent werre added to the test and blank wells of the 96-well plate respectively, then 25 μl of DTNB Solution was added it to the blank wells, and then 100 μl of Phosphate was added to the test wells and blank wells respectively. The plate was vibrated for 1 min and the absorbance at 405 nm (OD value) was determined after 5 min of standing.
Determination of Fe2+ Content
Intracellular Fe2+ content was determined using the Elabscience Cytosubstituted Ferrous Colourimetric Test Kit (Elabscience, China). The standards were configured and standard curve was plotted according to the instructions. One×106 cells was taked and 0.2ml buffer was added, mixed thoroughly and lysed on ice followed by, centrifuged at 15000rpm/10min. Eighty μl of supernatant was added to the 96-well plate, and 80 μl of colour development solution was added, and then incubated at 37 for 10min and measured the absorbance values (OD value) at 593nm. All experiments were performed independently three times.
Cell Proliferation Assay
Cell viability was analysed using CCK-8 reagent kit (MeilunBio, China). Cells were inoculated at 5 × 103/well (100 μl) in 96 plates and incubated for 24h. After 24h, 10 μl CCK-8 reagent was added to each well and incubation was continued for 2h. The absorbance values (OD values) at 450-490 nm were determined for each well, and all experiments were performed independently three times.
Apoptosis Assay
Since cellular lentiviral transfection contains fluorescence, the expression of apoptosis-related proteins (bcl-2, caspase-3) was detected using Western blot. Proteins were separated using SDSPAGE and transferred to a PVDF membrane. Primary antibodies bcl-2 (1:1000) and caspase-3 (1:2000) were incubated at 4°C for 12 hours. After removal of primary antibody, goat anti-rabbit secondary antibody (1:10000, Servicebio) was incubated for 12 hours at room temperature. Protein expression was detected by chemiluminescence, and grey scale values were processed and relative protein expression calculated using Image J software.
Western Blot
Total protein was extracted by adding RIPA lysis buffer (Servicebio) to the cells. Protein concentrations were determined using a BCA protein assay kit (Solarbio) and protein concentrations were adjusted so that they were consistent between the different groups. Proteins were separated using SDS-PAGE and transferred to a PVDF membrane (Billerica, USA). Primary antibodies TFRC (1:10000), ACSL4(1:10000), GPX4(1:5000), FTH1(1:2000), and GAPDH (1:500) were incubated for 12 h at 4°C. These antibodies were obtained from abcam, Cambridge, UK. After removing the primary antibody from the surface of the PVDF membrane using 1× TBST, the goat anti-rabbit secondary antibody (1:10000, Servicebio) was incubated for 12 h at room temperature. Protein expression was detected by chemiluminescence, and grey scale values were processed and relative protein expression calculated using Image J.
Statistical Analysis
Statistical analyses were performed using SPSS 21.0, and data were expressed as mean ± standard deviation(±SDs). Differences between groups were compared using t-test. P < 0.05 indicates statistically significant differences. All experiments were conducted independently at least three times.
Results
Microarray Determination of lncRNA Profiles and RT-qPCR Detection
To determine the expression of lncRNAs in HbH-CS disease, we collected blood samples from four pairs of HbH-CS disease patients and healthy individuals for microarray testing. The results showed that 5389 lncRNAs were differentially expressed (FC = 2, P < 0.05). Among them, 1393 lncRNAs were up-regulated and 3996 lncRNAs were down-regulated (Figure 1A-1C). The differentially expressed lncRNAs were also detected using RT-qPCR, which showed that SNHG6 was significantly upregulated in patients with HbH-CS disease (P < 0.05) (Figure 1D).
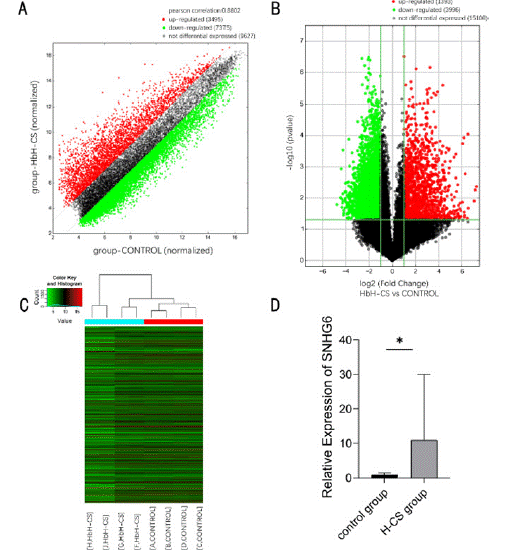
Figure 1: Comparison of lncRNA expression profiles between patients with HbH-CS disease and healthy individuals. (A) Scatter plot showing the distribution of
lncRNA expression. (B) Volcano plot showing differential expression of lncRNAs. (C) Clustered heat map showing differential expression of lncRNAs in HbH-CS
disease patients and healthy individuals. (D) RT-qPCR was used to detect the relative expression levels of SNHG6 in samples from HbH-CS disease patients and
healthy individuals, N = 15, *P < 0.05.
CeRNA Network Construction and Characterisation
The target miRNA of SNHG6 was predicted using the online tool Starbase and intersected with the original microarray data (Figure 2A). The related target miRNAs were detected using RT-qPCR, which showed that miR-579-3p was significantly downregulated in patients with HbH-CS disease (P < 0.05) (Figure 2B). Then the target mRNA of miR-579-3p was predicted using the online tool Starbase and crossed with the original microarray data (Figure 2C), while the target gene TFRC was identified based on the functional relevance of the protein. The expression of TFRC was detected using RT-qPCR, and the results showed that TFRC was significantly up-regulated in patients with HbH-CS disease (P < 0.01) (Figure 2D and 2E). Finally, SNHG6/miR- 579-3p/TFRC was identified as the ceRNA network for this study.
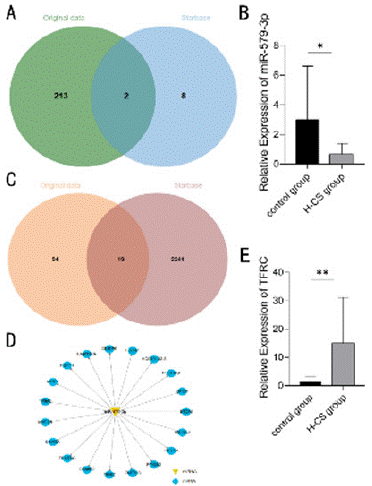
Figure 2: Bioinformatics analysis. (A) Venn diagram showing the pooling
of miRNAs in the microarray raw data with the online tool Starbase. (B)
RT-qPCR detection of the relative expression levels of miR-579-3p in
samples from patients with HbH-CS disease and healthy individuals, N = 15,
*P < 0.05. (C) Venn diagram showing the pooling of microarray raw data with
mRNAs in the online tool Starbase. (D) Predictive map of miR-579-3p target
genes. Yellow triangular nodes, miR-579-3p; blue circular nodes, mRNA.
(E) RT-qPCR was used to detect the relative expression levels of TFRC
in samples from patients with HbH-CS disease versus those from healthy
individuals, N = 15, **P < 0.01.
Dual Luciferase Reporter Gene Test
Bioinformatics analysis showed that miR-579-3p was able to bind to SNHG6 3‘UTR and TFRC 3’UTR, and the targeting relationship was verified using a dual luciferase reporter gene assay. The results showed that luciferase activity was significantly reduced after co-transfection of wild-type SNHG6 with miR-579-3p mimic, whereas the inhibition of luciferase activity was not obvisous after co-transfection of mutant SNHG6 with miR-579-3p mimic, which siggests that miR-579-3p was able to specifically bind to the SNHG6 3' UTR (P < 0.001) (Figure 3A and 3B). Similarly experiments demonstrated that miR-579-3p was able to specifically bind to the TFRC 3' UTR (P < 0.001) (Figure 3C and 3D). Targeting relationship in the SNHG6/miR-579-3p/TFRC network was existed.
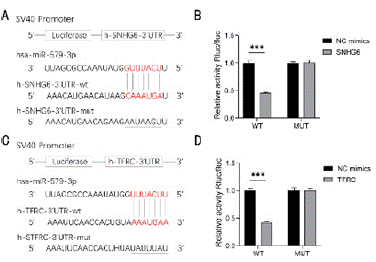
Figure 3: Dual luciferase reporter gene assay. (A) SNHG6 binding sites
to miR-579-3p. (B) Dual luciferase reporter gene assay for SNHG6-3’UTR
interaction with miR-579-3p, ***P < 0.001. (C) Binding site of miR-579-
3p with TFRC. (D) Dual luciferase reporter gene detection of miR-579-3p
interacting with TFRC-3’UTR, ***P < 0.001.
Expression of SNHG6, miR-579-3p, and TFRC in K562 Cells
To further validate the expression of the relevant genes in K562 cells, we constructed SNHG6 down-regulated cell lines and SNHG6 and miR-579-3p down-regulated co-transfected cell lines.
The subgroups were sh-SNHG6 NC group, sh-SNHG6 group, sh- SNHG6+inhibitor NC group, and sh-SNHG6+miR-579-3p inhibitor group. NC was the negative control, which was the interfering sequence of SNHG6 and miR-579-3p mimics. RT-qPCR results showed that miR-579-3p expression level of sh-SNHG6 +inhibitor NC group and the sh- SNHG6 group was significantly higher than the sh-SNHG6 NC group, while TFRC expression level of sh-SNHG6 +inhibitor NC group was significantly lower than the sh-SNHG6 NC group (P < 0.001 and P < 0.01) (Figure 4A and 4C). In addition, sh-SNHG6+miR-579-3p inhibitor group had a significantly higher SNHG6 expression level and TFRC expression level (P < 0.05 and P < 0.01) (Figure 4B).
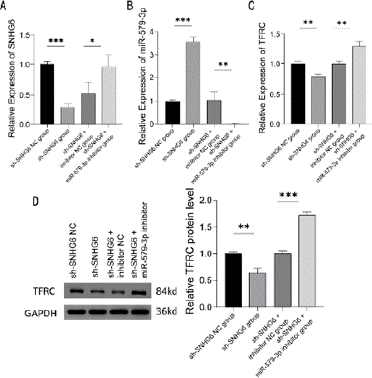
Figure 4: Expression of SNHG6, miR-579-3p, and TFRC. (A-C) RT-qPCR
detection of the expression levels of SNHG6 (A), miR-579-3p (B), and TFRC
(C) in K562 cells transfected with control, SNHG6 inhibitor, SNHG6 and miR-
579-3p inhibitor, N = 3, *P < 0.05, **P < 0.01, ***P < 0.001. (D) Western
blot analysis of protein expression levels of TFRC in K562 cells transfected
with control, SNHG6 inhibitor, SNHG6 and miR-579-3p inhibitor, N = 3, **P <
0.01, ***P < 0.001. Results are shown as mean ± SD from three independent
experiments.
Western blot results showed that TFRC protein expression level was significantly lower in sh-SNHG6 group and TFRC protein expression level was significantly higher in sh-SNHG6 +miR-579- 3p inhibitor group compared to the sh-SNHG6 NC group and sh- SNHG6 +inhibitor NC group (P < 0.01 and P < 0.001) (Figure 4D).
Effect of SNHG6/miR-579-3p on the Level of Oxidative Stress
In order to explore the effects of SNHG6 and miR-579-3p on the level of cellular oxidative stress, we determined the content of MDA and GSH in stably transfected cell lines. The results showed that the MDA content was significantly lower and the GSH content was significantly higher in the sh-SNHG6 group compared to the sh- SNHG6 NC group and sh-SNHG6 +inhibitor NC group (P < 0.05); while the MDA content had a significantly higher level and GSH content had a significantly lower level in the sh-SNHG6 +miR-579-3p inhibitor group (P < 0.05) (Figure 5A and 5B).
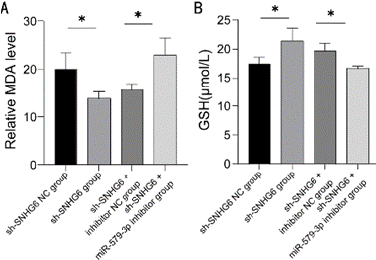
Figure 5: Detection of lipid oxidation in K562 cells. (A) Detection of MDA
content in K562 cells transfected with control, SNHG6 inhibitor, SNHG6
and miR-579-3p inhibitor, N = 3, *P < 0.05. (B) Detection of GSH content in
K562 cells transfected with control, SNHG6 inhibitor, SNHG6 and miR-579-
3p inhibitor, N = 3, *P < 0.05. Results are shown as mean ± SD from three
independent experiments.
Effect of SNHG6/miR-579-3p on Fe2+ Levels
The results of intracellular Fe2+ content measurement showed that Fe2+ content was significantly lower in the sh-SNHG6 group and higher in the sh-SNHG6+miR-579-3p inhibitor group, compared to the sh-SNHG6 NC group and sh-SNHG6+inhibitor NC group (P < 0.01 and P < 0.001) (Figure 6).
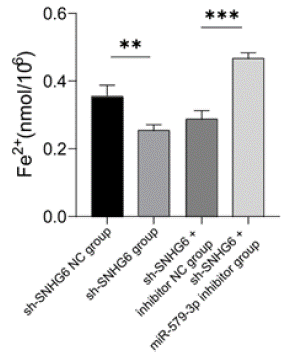
Figure 6: Detection of Fe2+ content in K562 cells. Detection of Fe2+ content in
K562 cells transfected with control, SNHG6 inhibitor, SNHG6 and miR-579-
3p inhibitor, N = 3, **P < 0.01, ***P < 0.001. Results are shown as mean ± SD
from three independent experiments.
Effect of SNHG6/miR-579-3p on Cell Proliferation Apoptosis
The results showed that the proliferation ability of cells in sh- SNHG6 group was significantly enhanced at 24h, and the proliferation ability of cells in sh-SNHG6+miR-579-3p inhibitor group was significantly inhibited at 12h compared to sh-SNHG6 NC group and sh-SNHG6+inhibitor NC group (P < 0.01) (Figure 7A and 7B).
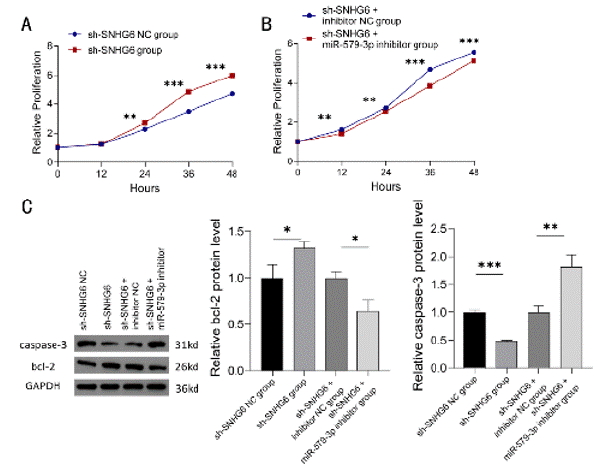
Figure 7: Detection of K562 cell proliferation and apoptosis. (A, B) Proliferation of K562 cells transfected with control, SNHG6 inhibitor, SNHG6 and miR-579-3p
inhibitor was detected by CCK8, N = 3, **P < 0.01, ***P < 0.001. (C) Western blot assay was performed to detect the protein expression levels of apoptosisrelated
genes bcl-2 and caspase-3 in K562 cells transfected with control, SNHG6 inhibitor, SNHG6 and miR-579-3p inhibitor. N = 3, *P < 0.05, **P < 0.01,
***P < 0.001. Results are shown as mean ± SD from three independent experiments.
Western blot detection of apoptosis-related protein expression showed that, compared with sh-SNHG6 NC group and sh- SNHG6+inhibitor NC group, the expression level of bcl-2 protein in the sh-SNHG6 group was significantly higher, while the expression level of caspase-3 protein was significantly decreased (P < 0.05 and P < 0.001); moreover, bcl-2 protein expression level was significantly decreased and caspase-3 protein expression level was significantly increased in sh-SNHG6+miR-579-3p inhibitor group compared to the NC group (P < 0.05 and P < 0.01) (Figure 7C).
Effect of SNHG6/miR-579-3p on the Expression of Iron Death-related Proteins
The expression of iron death-related proteins was detected by Western blot. The results showed that compared with the sh-SNHG6 NC group and sh-SNHG6+inhibitor NC group, the expression level of ACSL4 protein was significantly lower (P < 0.001), the expression level of GPX4 protein was significantly higher (P < 0.01), and the FTH1 protein expression levels were significantly higher in the sh- SNHG6 group (P < 0.01); while the ACSL4 protein expression levels were significantly higher (P < 0.01), GPX4 protein expression levels were significantly lower (P < 0.001), and FTH1 protein expression levels were significantly lower (P < 0.001) in the sh-SNHG6+miR- 579-3p inhibitor group (Figure 8A-8C).
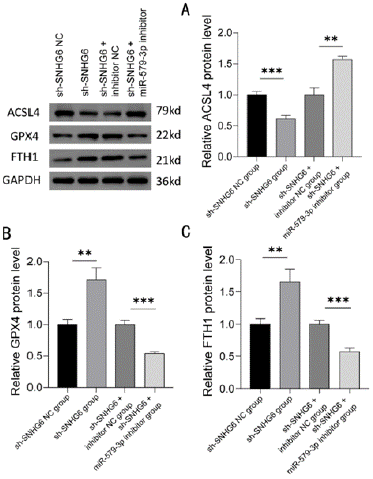
Figure 8: Western blot analysis of ACSL4, GPX4 and FTH1 expression.
Protein expression levels of iron death-related genes ACSL4 (A), GPX4
(B), and FTH1 (C) in K562 cells transfected with control, SNHG6 inhibitor,
SNHG6 and miR-579-3p inhibitor were detected by Western blot, N = 3, **P <
0.01, ***P < 0.001. Results are shown as mean ± SD from three independent
experiments.
Discussion
In this study, we investigated lncRNA SNHG6 as an endogenous competitive RNA to bind to miR-579-3p thereby deregulating the inhibitory effect of miR-579-3p on TFRC, and explored the effects of SNHG6/miR-579-3p/TFRC on HbH-CS disease in terms of oxidative stress, Fe2+, apoptosis, and cell proliferation and iron death.
HbH-CS is one of the common thalassaemia syndromes, with a marked decrease in haemoglobin and severe clinical manifestations, including hepatosplenomegaly, growth retardation or transfusiondependent haemoglobinopathy in severe cases [16,17]. HbH-CS disease is due to the non-deletion gene mutation that reduces the production of a-globin chains and leads to ineffective erythropoiesis, and the imbalance of a- globin chains and Β-globin chains can form unstable haemoglobin tetramers, and the decomposition of these tetramers releases a large amount of free iron, reactive oxygen species and so on to further promote erythropoietic cell death, which aggravate the anaemia [7,8]. Transcriptional and epigenetic mechanisms are important intrinsic factors regulating erythropoiesis, and lncRNAs play an important role in transcriptional regulation. It has now been demonstrated that lncRNAs play an important role in the transcription of erythroid lineage genes and are involved in the regulation of the terminal maturation stage during the proliferation and differentiation of erythroid cells [11,14]. Previous studies have demonstrated that lncRNAs play an important role in the pathogenesis of haematopoietic and blood disorders, e.g. lincRNAEPS promotes apoptosis in mouse erythroid precursors [18]; multiple erythroid-restricted expression of lncRNA transcripts is able to target key erythroid transcription factors [19]; and lincRNA SRA promotes the expression of the erythroid lineage in primary human erythroid progenitor cells program transcriptome expression and expression of red lineage markers, among others [20]. With the identification of lncRNAs in hematopoiesis, it can help us to better understand the role of lncRNAs in blood cells and may be a new marker for diagnosis, prognosis and prediction of response to therapy in diseases [21]. However, most studies of lncRNAs in thalassaemia have focused on Β-thalassaemia, for example, NR-001589 can activate HBE1 and haematopoietic lineage-inducing factor epitopes thereby increasing the expression of HbF [14]; MALAT1 may be associated with the phenotype and comorbidities of Β-thalassaemia, and MIAT may be correlated with the severity of Β-thalassaemia [12]. In a-thalassaemia, lncRNA-aGT is able to promote the activation of the a- globin chains adult gene [22]. These existing experimental results have focused on Β-thalassaemia, while the differential expression and mechanism of action of lncRNA in HbH-CS disease have rarely been investigated. Therefore, in order to find out whether there is differential expression of lncRNA in HbH-CS disease and to further understand the related mechanism, we collected peripheral blood of patients with HbH-CS disease and healthy individuals for microarray sequencing, to identify the role of lncRNA in the differential expression of lncRNA in HbHCS disease. A total of 5389 lncRNAs were found to be differentially expressed, of which 1393 lncRNAs were upregulated and 3996 lncRNAs were downregulated. To further understand the relevant roles and mechanisms played by lncRNAs, we screened the microarray raw data and identified them by RT-qPCR, which showed that the expression of SNHG6 was significantly elevated in HbH-CS disease. It has been demonstrated that SNHG6 may be involved in regulating the process of haematopoietic differentiation, and knockdown of SNHG6 was able to cause differences in the expression of erythrocyte-specific genes (e.g. haemoglobin subunits) and platelet-specific genes [23]. Therefore, we speculate that SNHG6 may play an important role in erythropoiesis and haemoglobin production in HbH-CS disease.
We identified the existence of a targeting relationship between SNHG6 and miR-579-3p by the online tool Starbase prediction as well as a dual luciferase reporter gene assay, and identified that miR- 579-3p expression was significantly reduced in HbH-CS disease by RT-qPCR. It has been shown that miR-579-3p plays an important role in cell proliferation and migration, and is able to regulate the iron death process through interaction with SQLE [24]. We again identified the existence of a targeting relationship between miR-579- 3p and TFRC by the online tool Starbase prediction as well as a dual luciferase reporter gene assay, and identified a significantly elevated expression of TFRC in HbH-CS disease by RT-qPCR. It has been shown that TFRC is specifically expressed in erythroid precursors and decreases with erythroid differentiation, and is one of the major factors in iron uptake during erythroid growth and development [25]. Iron is required for the synthesis of haemoglobin during erythrocyte growth and development, and up-regulation of TFRC and porphyrin synthesis genes in erythrocytes during the final stages of erythrocyte differentiation can increase iron uptake and haemoglobin production [26]. Pathological reduction of TFRC expression in erythrocytes results in microcytic anaemia [27,28]. Patients with HbH-CS have severe anaemia, with predominantly ineffective erythropoiesis, and up-regulation of TFRC promotes the uptake of iron into the cells, thereby increasing erythropoiesis. Iron is an essential nutrient for biological processes such as oxygen transport, protein synthesis, DNA replication, and oxidative phosphorylation [29], and its electrongaining and electron-losing properties also favour the production of reactive oxygen species (ROS), so that intracellular iron levels need to be strictly controlled [30,31]. TFRC is able to translocate extracellular iron intracellularly, and up-regulation of TFRC increases iron uptake as well as the pool of unstable redox-activated iron, which leads to iron death [32,33]. Upregulation of TFRC in patients with HbH-CS disease increases intracellular iron content, with some risk of causing cellular iron death.
To explore the role of SNHG6/miR-579-3p/TFRC, we further investigated it in in vitro experiments. K562 is a chronic myeloid leukaemia cell line with an embryonic/fetal globin gene expression pattern that is induced to differentiate into an erythroid cell line. Therefore, we chose K562 cells for in vitro experiments. The experiments showed that down-regulation of SNHG6 would upregulate the expression level of miR-579-3p, down-regulate the expression level of TFRC, reduce the level of cellular oxidative stress, reduce the level of intracellular Fe2+, promote cell proliferation, inhibit apoptosis, up-regulate the expression of GPX4 and FTH1, and down-regulate the expression of ACSL4; Down-regulation of miR- 579-3p was able to compensate for the effect of down-regulation of SNHG6; up-regulation of the expression level of TFRC, increased the level of cellular oxidative stress, increased the level of intracellular Fe2+, inhibited cellular proliferation, and promoted apoptosis, down-regulation of the expression of GPX4 and FTH1, and up-regulation of the expression of ACSL4.
These experiments demonstrated that down-regulation of SNHG6 was able to reduce TFRC expression thereby decreasing the transfer of extracellular iron into the cell, decreasing intracellular Fe2+ levels, decreasing the level of intracellular oxidative stress, and decreasing the pool of unstable redox iron. The down-regulation of TFRC expression increased the expression of GPX4, FTH1, and the up-regulation of GPX4 expression increased the reduction of various oxidative substrates thereby protecting the cells from damage caused by oxidative stress [34,35], and up-regulation of FTH1 expression increases storage and detoxification of excess intracellular iron [36], thereby reducing cell death due to increased intracellular iron and oxidative stress. In contrast, downregulation of TFRC expression decreases ACSL4 expression, which decreases the sensitivity of cells to iron death and inhibits the activation of cellular iron death [37,38].
Conclusions
In conclusion, we identified differentially expressed lncRNAs in HbH-CS disease. Our results suggest that SNHG6, miR-579-3p and TFRC can constitute a ceRNA network, and that down-regulation of SNHG6 reduces the level of cellular oxidative stress and inhibits the activation of iron death. Thus, we have preliminarily elucidated the mechanism of SNHG6/miR-579-3p/TFRC network action on HbHCS disease, further deepening our understanding of HbH-CS disease.
Author Contributions
Chunjiang Zhu was the guarantor and designed the study; Xinyu Wang performed the experiments; Yulan Li coordinated blood sample collection and bioinformatics anlysis; Limin Mo interpreted the results; Guanjun Chen contributed to bioinformatics analysis; Xinyu Wang, Jun Li and Malik KASHIF drafted the initial manuscript; Wei Hou provided the laboratory and experimental apparatus and materials; Chunjiang Zhu revised the article critically for important intellectual content. All authors approved the final manuscript and agreed to submit for publication.
Acknowledgments
We thank Dr. Zhang XH for the assistance in specimen collection.
Funding
This study was supported by the Guangxi Medical and health key discipline construction project, the National Natural Science Foundation of China (no.82060037), and the Guangxi Ba Gui Scholars Special Project.
Data Availability Statement
All raw data of this study are available from the corresponding author by request.
References
- Tesio N, Bauer DE. Molecular Basis and Genetic Modifiers of Thalassemia. Hematology/Oncology Clinics of North America. 2023; 37: 273-299.
- Xiong F, Sun M, Zhang X, et al. Molecular epidemiological survey of haemoglobinopathies in the Guangxi Zhuang Autonomous Region of southern China. Clin Genet. 2010; 78: 139-148.
- Chen P, Lin WX, Li SQ. THALASSEMIA in ASIA 2021: Thalassemia in Guangxi Province, People’s Republic of China. Hemoglobin. 2022; 46: 33-35.
- Higgs DR, Gibbons RJ. The Molecular Basis of a-Thalassemia: A Model for Understanding Human Molecular Genetics. Hematology/Oncology Clinics of North America. 2010; 24: 1033-1054.
- Chui DHK, Fucharoen S, Chan V. Hemoglobin H disease: not necessarily a benign disorder. Blood. 2003; 101: 791-800.
- Cheng HL, Liu RR. Advances in the treatment of haemoglobin H disease. Chinese Journal of Comparative Medicine. 2019; 29: 110-115.
- Lal A, Viprakasit V, Vichinsky E, Lai Y, Lu MY, Kattamis A. Disease burden, management strategies, and unmet needs in a-thalassemia due to hemoglobin H disease. Am J Hematol. 2024; 99: 2164-2177.
- Lal A, Goldrich ML, Haines DA, Azimi M, Singer ST, Vichinsky EP. Heterogeneity of Hemoglobin H Disease in Childhood. New England Journal of Medicine. 2011; 364: 710-718.
- Marchese FP, Huarte M. Long non-coding RNAs and chromatin modifiers. Epigenetics. 2013; 9: 21-26.
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi Pier P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. 2011; 146: 353-358.
- Siatecka M, Kulczynska K. A regulatory function of long non-coding RNAs in red blood cell development. Acta Biochimica Polonica. 2017; 63: 675-680.
- Fakhr-Eldeen A, Toraih EA, Fawzy MS. Long non-coding RNAs MALAT1, MIAT and ANRIL gene expression profiles in beta-thalassemia patients: a cross-sectional analysis. Hematology. 2019; 24: 308-317.
- Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes to Cells. 2011; 16: 479-490.
- Lai K, Jia S, Yu S, Luo J, He Y. Genome-wide analysis of aberrantly expressed lncRNAs and miRNAs with associated co-expression and ceRNA networks in Β-thalassemia and hereditary persistence of fetal hemoglobin. Oncotarget. 2017; 8: 49931-49943.
- Starbase. 2023.
- Fucharoen S, Viprakasit V. Hb H disease: clinical course and disease modifiers. Hematology. 2009; 2009: 26-34.
- Singer ST, Kim HY, Olivieri NF, et al. Hemoglobin H-constant spring in North America: An alpha thalassemia with frequent complications. American Journal of Hematology. 2009; 84: 759-761.
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated antiapoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011; 25: 2573-2578.
- Alvarez -Dominguez JR, Hu W, Yuan B, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014; 123: 570-581.
- Sawaengdee W, Cui K, Zhao K, Hongeng S, Fucharoen S, Wongtrakoongate P. Genome-Wide Transcriptional Regulation of the Long Non-coding RNA Steroid Receptor RNA Activator in Human Erythroblasts. Front Genet. 2020; 11: 850.
- Nobili L, Lionetti M, Neri A. Long non-coding RNAs in normal and malignant hematopoiesis. Oncotarget. 2016; 7: 50666-50681.
- Arriaga-Canon C, Fonseca-Guzmán Y, Valdes-Quezada C, Arzate-Mejía R, Guerrero G, Recillas-Targa F. A long non-coding RNA promotes full activation of adult gene expression in the chicken a-globin domain. Epigenetics. 2014; 9: 173-181.
- Hazan JM, Amador R, Ali-Nasser T, et al. Integration of transcription regulation and functional genomic data reveals lncRNA SNHG6’s role in hematopoietic differentiation and leukemia. Journal of Biomedical Science. 2024; 31: 27.
- Li Peili, Wen Lifang. Study on the malignant behaviour of cervical cancer HeLa cells by miR-579-3p targeting squalene monooxygenase to regulate iron death. Chinese Journal of Clinical Pharmacology. 2023; 39: 1894-1898.
- Iacopetta BJ, Morgan EH, Yeoh GC. Transferrin receptors and iron uptake during erythroid cell development. Biochim Biophys Acta. 1982; 687: 204- 210.
- Chen M, Zhang Y, Jiang K, et al. Grab regulates transferrin receptor recycling and iron uptake in developing erythroblasts. Blood. 2022; 140: 1145-1155.
- Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999; 21: 396-399.
- Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005; 106: 1084-1091.
- Arredondo M, Núñez MT. Iron and copper metabolism. Mol Aspects Med. 2005; 26: 313-327.
- Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001; 33: 940-959.
- Weinberg ED. The hazards of iron loading. Metallomics. 2010; 2: 732-740.
- Feng H, Schorpp K, Jin J, et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Reports. 2020; 30: 3411-3423.e3417.
- Yang WS, Stockwell BR. Synthetic Lethal Screening Identifies Compounds Activating Iron- Dependent, Nonapoptotic Cell Death in Oncogenic-RASHarboring Cancer Cells. Chemistry & Biology. 2008; 15: 234-245.
- Yant LJ, Ran Q, Rao L, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003; 34: 496-502.
- Weaver K, Skouta R. The Selenoprotein Glutathione Peroxidase 4: From Molecular Mechanisms to Novel Therapeutic Opportunities. Biomedicines. 2022; 10: 891.
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996; 1275: 161-203.
- Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016; 478: 1338-1343.
- Doll S, Proneth B, Tyurina YY, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017; 13: 91-98.