
Research Article
Austin Food Sci. 2021; 6(1): 1042.
Functionally Active Microbiome and Physicochemical Properties of Milk and Sugary Water Kefir from Brazil
Villanoeva CNB1#, Rios DL2#, Alvarenga RL3, Acurcio LB1, Sandes SHC2, Nunes AC2, Nicoli JR1 and Neumann E1*
¹Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Brazil
²Departamento de Genética, Ecologia e Evolução, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Brazil
³Laboratório de Bromatologia, Faculdade de Farmácia, Universidade Federal de Minas Gerais, Brazil #Equal Contribution to the Execution of Experiments
*Corresponding author: Neumann E, Universidade Federal de Minas Gerais, Campus Pampulha, Av. Antônio Carlos, Brazil
Received: February 09, 2021; Accepted: March 09, 2021; Published: March 16, 2021
Abstract
In Brazil, milk kefir, made of milk kefir grains, has identity features defined by Brazilian regulatory agencies but sugary water kefir, made of water kefir grains, has no definition of microbiological and physicochemical standards. We evaluated the microstructure of Brazilian milk and water kefir grains, the Transcriptionally Active Microbiome (TAM) of kefir beverages made of them, and the effect of fermentation and storage period (28 days at 10oC) over microbiological and physicochemical features of these beverages. Milk and water grains are very different between them and similar to other kefir grains worldwide, macroscopically and microscopically. The genus Leuconostoc, with the species L. mesenteroides, was most frequent in the microbiome of milk kefir while the Oenococcus genus was most frequently seen in sugary water kefir, with the species O. kitaharae. The genera Saccharomyces and Torulaspora, with the species S. cerevisiae and T. delbrueckii, were most recurrent in the microbiome of sugary water kefir, while Pichia and Yarrowia were more abundant in milk kefir, with the species P. fermentans and Y. lipolytica. Microbiological and physicochemical parameters of milk kefir were in concordance with features defined by Brazilian legislation. None of the parameters was altered by cold storage for 28 days. Our results reinforce some Brazilian identity requirements for milk kefir and allow us to suggest the inclusion of new ones that are not defined yet. Regarding sugary water kefir, some microbiological and physicochemical parameters are similar to milk kefir during the same storage period, although with a quite different functional microbiome.
Keywords: Brazilian Kefir grains; Brazilian kefir beverages; Sugary water kefir identity; Functional microbiome
Introduction
Kefir is a kind of fermented milk originated from the Caucasian mountains and dispersed worldwide. It is obtained from the fermentation by starter microorganisms present in typical grains. Kefir grains possess a microbiota composed of an association between Lactic Acid Bacteria (LAB), Acetic Acid Bacteria (AAB), and yeast, entrapped by an Exopolysaccharide (EPS) matrix [1-4]. When inoculated in milk, the grains’ microbiota produces lactic and acetic acid, ethanol, CO2, and aromatic compounds, leading to pH reduction with protein precipitation. These microorganisms that give a distinct character to the drink should be viable and abundant until the predetermined expiry date [5-8]. Sugary water kefir is usually produced in water added with brown sugar (around 5% w/v) or fruit juices, by addition of water kefir grains. Fermentation from both kefir occurs for 24 to 48 hours at room temperature, producing turbid, sparkling, acid, and slightly alcoholic beverages [9-11].
Kefir has been consumed due to the health benefits produced by potentially probiotic microorganisms isolated from kefir grains and beverages [12,13]. In addition, EPS production from LAB is a significant feature due to its rheological improvements to the beverages and its potential functional properties [14,15].
In Brazil, there is an incipient industrial production of kefir. Kefir is often produced at domestic level by using two types of kefir grains that circulate the country, milk kefir grains used to produce the traditional fermented milk, and water kefir grains used to prepare a watery fermented drink with brown sugar. The geographic origin of grains, with different climate conditions, the grains subculture methods and the substrate used for fermentation may result in alterations of the beverage’s characteristics. Indeed, there are few data available regarding microbial composition and scientific literature concerning quality and physicochemical standards for the different kefir beverages produced in Brazil [2-4,16-19].
Transcriptomics deals with the complete set of RNA transcripts produced by the microbial cells in a specific time or place using high-throughput NGS technologies called RNA-Seq [20]. Therefore, a more accurate estimative of the abundance of active bacteria and yeasts of kefir community could be achieved seeking mRNA transcripts of housekeeping and ribosomal protein genes, generating a Transcriptionally Active Microbiome (TAM).
Thus, this work aimed to assess the functional microbiome of different milk and water Brazilian kefir and evaluate the effect of fermentation and cold storage period on the microbiological and physicochemical characteristics of those beverages. These results could contribute to define a true identity for both types of beverages produced in Brazil.
Materials and Methods
Kefir grains
Kefir grains used in this study were cultivated in two different food matrixes: milk and water (with brown sugar), according to its original propagation matrix. The water kefir grains were provided from different domestic environments from Brazilian cities of Belo Horizonte (KABH), Curitiba (KACU), and Salvador (KASA). Milk grains were from Curitiba (KLCU), Salvador (KLSA), and Divinópolis (KLDI). Water and milk grains from Viçosa (KAVI and KLVI, respectively) were from the Fermented Dairy Products Laboratory from the Federal University of Viçosa (UFV). All grains were kept at -86°C until their usage.
Electron microscopy of kefir grains
Approximately 0.5g of milk and water kefir grains from Curitiba and Salvador were prepared according to procedures for electron microscopy (scanning and transmission) [22], which were conducted at Microscopy Center from UFMG. Preparation for Scanning (SEM) and Transmission (TEM) electronic microscopic was realized through the Osmium-Tannin-Osmium (OTO) method. SEM samples were analyzed in an electronic scanning microscope FEG - Quanta 200 (Fei Tecnai, Oregon, USA). TEM samples were cut by microtome and analyzed in an electronic transmission microscope - Tecnai Spirit Biotwin G2-12 (Fei Tecnai, Oregon, USA).
Kefir production
Milk kefir was prepared with reconstituted skim milk powder (10% w/v) and sugary water kefir with brown sugar solution (5% w/v). After sterilization, both substrates received a 3% w/v of specific grain inoculum, and they were incubated for 24h at 25°C, followed by maintenance at 10±2°C for 24h, sieving grains and the fermented beverages stored at 10±2°C for 28 days. All experiments were done with four repetitions.
Microbiological and physicochemical properties of kefirfermented beverages
Microbiological properties were evaluated through LAB enumeration in De Man, Rogosa and Sharpe (MRS, Acumedia, Lansing) and yeast count in Potato Dextrose Agar (PDA, Acumedia). The physicochemical analysis included pH, titratable acidity, fat, protein and lactose measurements [22]. The analysis was conducted, always duplicated, at 1, 2, 7 and 28 days post-inoculation of substrates with the grains.
Identification of the transcriptionally active microorganisms in kefir-fermented beverages by transcriptomic analysis (RNA-seq)
One milliliter of kefir-fermented milk or 40 mL of water kefir was centrifuged for 10min at 10,000xg, the cell pellets were transferred to microtubes containing 0.3g of zirconium beads, ruptured in the FastPrep-24 instrument (MP Biomedicals), and total RNA was extracted using the RNAeasy mini kit (Qiagen), according to the manufacturer’s recommendations. The extracted RNA was reversedtranscripted to cDNA do build libraries for NGS sequencing. The samples were divided into two parts, one destined to analyze the bacteria and the other the study of yeasts. The bacterial sample was treated with the Ribo-Zero rRNA removal kit, and the yeast sample was enriched with the capture of mRNAs by the poly-A tail, all the procedures according to the manufacturer’s recommendations (Illumina).
The cDNA libraries were elaborated according to the RNA Sample sequencing protocol from Illumina, and sequencing by bridging PCR in MiSeq sequencer, as stated by the manufacturer (Illumina).
MiSeq reagent kit v3 (600-cycle) was used to enable the highest output of sequenced information (15Gb, 2x300 bp, up to 25 million reads).
Analysis of bioinformatics
The computational issues were developed in the servers Sagarana and Truta, located in the Laboratories of Informatics of the ICB/ UFMG and Fiocruz/MG, in GNU Linux/Debian operating system. Some small computational algorithms were developed throughout the project. These scripts were made in Python programming language. The computational strategy used was multithreading, aiming to increase performance and reduce the processing time associated with the programs used.
To perform the first stage of pipeline development for the RNAseq analysis, Trimmomatic and FastQC were used for pre-processing and quality analysis of the reads. Then, FASTQ-Join [23] merged the sequences forward and reverse to form consensus sequences that were aligned via MegaBLAST with NCBI NT database (nucleotide sequences), by the software HS-BLASTN [24]. In the third step of the pipeline for RNA-seq analysis, assembly in contigs and functional annotation of reads were performed using Trinity software [25], Transdecoder [26], AC-DIAMOND and STAR software. The Transdecoder identifies which contigs are mRNA and what possible ORFs. The AC-DIAMOND aligns by BLASTx the annotated contigs as mRNA against the NCBI NR database (non-redundant protein sequences) and UEKO-UniRef Enriched KEGG Orthology [27]. Finally, STAR software aligns the reads again against the contigs annotated as mRNA for quantifying the gene expression.
The transcriptionally active microbiome of Bacteria by multilocus sequence analysis (bTAM) and yeast by rRNA ITS sequence analysis (fTAM)
BLASTx searched contigs related to the housekeeping and ribosomal protein genes in the UniProt revised protein database for MLSA and rMLSA analysis [28]. Seventy-eight housekeeping markers related to the RNA polymerase core subunits and sigma factors, RNA polymerase-associated proteins, transcription elongation and termination factors, DNA replication initiation, elongation and termination factors, and DNA topoisomerases were chosen for MLSA; and seventy-five genes related to the ribosome-associated proteins and protein translation initiation, elongation, and release factors were selected for rMLSA (Supplementary Table S1).
For the identification of yeasts, it was used a dataset with all ITS (Fungal Internal Transcribed Spacer RNA) sequences available in the RefSeq Targeted Loci Project-Bio Project, of NCBI.
Statistical analysis
Statistical analysis was conducted at SAS software 9.2 (SAS Institute Inc., Cary, NC, USA), at a 5% significance level. The effects of storage conditions on the microbiological and physicochemical characteristics of kefir were determined by Analysis of Variance (ANOVA) and, if necessary, by the Tukey test.
Results and Discussion
Microstructure of Brazilian kefir grains
Brazilian milk kefir grains studied herein were in general small, round, with an irregular shape, white or yellow, similar to cauliflower pieces. Water kefir grains had a consistent gelatinous aspect, yellow and translucid, with irregular shape and size. Both types of grains are very different between them and similar to other kefir grains worldwide [2-4,29-31].
Scanning Electronic Microscopic (SEM) of two Brazilian milk kefir grains whose internal portions were obtained by cryogenic fracture showed, in both of them, presence of yeast at surface level (Figure 1A) and also at the internal part of the grain (Figure 1B). At the grains’ surface, it was possible to observe rod-shaped bacteria (Figure 1A-1), a large number of yeasts with short and long shape (Figure 1A-2), and granular material (Figure 1A-3), which has been described as clotted protein [2]. At the internal portion, bacilli and yeast, all involved by a fibrillar and porous structure to which microorganisms are attached to, can be observed as described by other authors [4,21,29,31].

Figure 1: Scanning Electronic Microscopic (SEM) of Brazilian milk kefir grains. Micrograph A: Outer surface of KLSA grains. 1-Bacillus, 2-Yeast, and 3-Granular
material (cotted protein). Micrograph B: Inner part of fractured KLSA grains. Presence of bacteria (long and rods; bacilli in pairs) and yeast. 4-Fibrillar material:
EPS kefiran.
Brazilian water kefir grains presented bacteria and yeast adhered to a matrix that covers the whole grain surface (Figure 2A) and the internal surface (Figure 2B), as it was seen in milk grains. This matrix is smooth and poreless on the outside and spongy on the inside. Yeast shape was similar to the previously described for Brazilian milk kefir grains. Microbial density was lower than the observed in milk kefir grains, especially at the core of the grain. These characteristics was similar to the observed for others water grains studied worldwide [3,32].

Figure 2: Scanning Electronic Microscopic (SEM) of Brazilian sugary water kefir grains. Micrograph A: Outer surface of KASA grains. Micrograph B: Inner part of fractured KASA grains. 1-Bacillus, 2-Yeast (rod and elongated shapes), and 3-Polysaccharidic matrix.
The SEM findings were confirmed by Transmission Electron Microscopy (TEM) (Figure 3A). Microbiota from grains was active at the moment of fixation since it could be observed duplicating bacteria and budding yeasts (Figure 3B and 3C).
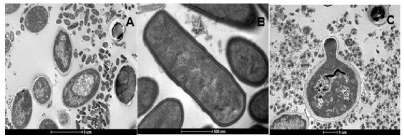
Figure 3: Transmission Electron Microscopy (TEM) of Brazilian kefir grains. Micrograph A: KLSA grain. Micrograph B and C: Metabolically active grains (presence
of bacteria in the process of duplication and budding yeasts, respectively.
Milk kefir microbiological and physicochemical characteristics
There was no significant difference in average viable counts of LAB and yeast in milk kefir during 28 days of cold storage (10°C) for almost all the beverages made with grains from different Brazil places (Table 1). The maintenance of at least 107CFU/ml of LAB and 104CFU/ml of Yeast for 28 days is in accordance with national and international standards for kefir [6,7]. Since LAB and yeast counts did not change significantly between the first and the second day of fermentation, it could be established 24h at 25°C as fermentation condition. In terms of industrial production, reducing the time of fermentation means reduced costs overall.
Microorganism
Sample
Time (Days)
P-Value
1
2
7
28
LAB
(Log CFU/mL)KLCU
7.00 ± 0.82a
7.58 ± 0.49a
7.67 ± 0.47a
7.18 ± 0.88a
0.489
KLDI
7.22 ± 1.12a
7.57 ± 1.00a
7.33 ± 1.22a
6.93 ± 0.95a
0.8677
KLSA
6.77 ± 0.37a
7.89 ± 0.88a
7.55 ± 1.00a
7.58 ± 0.87a
0.3009
KLVI
6.84 ± 0.66a
7.35 ± 0.93a
7.47 ± 1.33a
6.78 ± 0.79a
0.6691
Yeast
(Log CFU/mL)KLCU
5.41 ± 0.19b
5.97 ± 0.28a
6.05 ± 0.17a
6.25 ± 0.27a
0.0016
KLDI
5.55 ± 0.32b
6.22 ± 0.22a
6.23 ± 0.25a
6.36 ± 0.25a
0.0039
KLSA
5.78 ± 0.46a
6.12 ± 0.27a
6.19 ± 0.19a
6.13 ± 0.44a
0.4139
KLVI
5.70 ± 0.60a
6.00 ± 0.47a
6.38 ± 0.43a
6.42 ± 0.55a
0.2097
Different lowercase letters in the lines indicate a significant difference by the Tukey test (P <0.05). KLCU-Curitiba; KLDI-Divinópolis; KLSA-Salvador; KLVI-Viçosa.
Table 1: LAB and yeast counts of milk kefir produced with grains of different origins, over time (days) (n=4).
Beverages’ pH values ranged from 4.54 to 5.05, which is close to criteria used to stop the fermentation process of fermented milk [33], and did not significantly vary during 28 days of cold storage in all milk kefir. Rapid pH reduction is essential to alter the drink consistency [34,35]. The pH of the kefir also influences its microbiome, since it favours lactic acid bacteria and reduces most pathogenic bacteria [5,34]. Titratable acidity increased in all beverages during the cold storage. Acidity rise can be explained by the metabolic activity of acetic and lactic acid bacteria in kefir and lactose fermentation by some yeast strains [4].
As expected, lactose concentration reduced significantly (P<0.05) in all beverages along the storage time (Table 2). The lower lactose content is desirable considering that lactose intolerance affects, worldwide, a significant portion of the adult population, and fermented products, such as kefir, appears as a great alternative to dairy consumption [36,37]. Protein content did not change during fermentation or cold storage. Regarding fat content, our milk kefir can be classified as skim, as it was used milk with 0.5% of fat as substrate (Table 2).
Parameter
Sample
Time (Days)
P-Value
1
2
7
28
pH
KLCU
4.59 ± 0.43a
4.68 ± 0.44a
4.62 ± 0.30a
4.54 ± 0.26a
0.9443
KLDI
4.81 ± 0.27a
4.68 ± 0.30a
4.59 ± 0.26a
4.54 ± 0.19a
0.4948
KLSA
5.05 ± 0.12a
4.97 ± 0.14a
4.86 ± 0.15a
4.76 ± 0.20a
0.0922
KLVI
4.95 ± 0.19a
4.86 ± 0.23a
4.74 ± 0.16a
4.64 ± 0.22a
0.1975
Titratable aciditya
KLCU
0.74 ± 0.37a
0.83 ± 0.26a
1.02 ± 0.33a
1.30 ± 0.39a
0.1611
KLDI
0.70 ± 0.20b
0.85 ± 0.16ab
0.98 ± 0.24ab
1.19 ± 0.25a
0.0416
KLSA
0.59 ± 0.15c
0.70 ± 0.11bc
0.84 ± 0.03ab
1.02 ± 0.08a
0.0004
KLVI
0.61 ± 0.14c
0.75 ± 0.11bc
0.88 ± 0.10ab
1.07 ± 0.13a
0.0012
Lactoseb
KLCU
ND
3.57 ± 0.40a
3.30 ± 0.34a
2.77 ± 0.49a
0.0623
KLDI
ND
3.32 ± 0.47a
3.15 ± 0.32a
2.82 ± 0.28a
0.1966
KLSA
ND
3.61 ± 0.41a
3.44 ± 0.23ab
2.87 ± 0.42b
0.0444
KLVI
ND
3.55 ± 0.50a
3.29 ± 0.35a
2.90 ± 0.43a
0.148
Fatb
KLCU
ND
0.11 ± 0.03a
0.10 ± 0.01a
0.13 ± 0.01a
0.1792
KLDI
ND
0.11 ± 0.04a
0.11 ± 0.01a
0.12 ± 0.02a
0.9995
KLSA
ND
0.11 ± 0.04a
0.10 ± 0.01a
0.13 ± 0.01a
0.5332
KLVI
ND
0.11 ± 0.02a
0.11 ± 0.02a
0.12 ± 0.01a
0.2009
Proteinb
KLCU
ND
3.70 ± 0.09a
3.61 ± 0.08a
3.67 ± 0.22a
0.6778
KLDI
ND
3.67 ± 0.17a
3.79 ± 0.09a
3.74 ± 0.09a
0.4275
KLSA
ND
3.68 ± 0.07a
3.64 ± 0.13a
3.69 ± 0.12a
0.7602
KLVI
ND
3.73 ± 0.13a
3.67 ± 0.08a
3.60 ± 0.16a
0.3938
aTitratable acidity expressed as mg lactic acid/mL; bLactose, fat, and protein expressed as g/100g. Different lowercase letters in the lines indicate a significant difference by the Tukey test (P <0.05). KLCU-Curitiba; KLDI-Divinópolis; KLSA-Salvador; KLVI-Viçosa.
Table 2: Physicochemical features of milk kefir produced with grains of different origins, over time (days) (n=4).
Fermented milk tested herein attended national [6] and international [7] requirements after 48h of fermentation (Table 3). Since there is no standard criteria on pH and lactose content, we suggest 4.6 for pH and 3.5g/100g of lactose, considering the average values for milk kefir analyzed.
Parameter
KLCU
KLDI
KLSA
KLVI
RTIQ
Brasil (2007)Codex Alimentarius
(2018)LAB (Log CFU/mL)
3.80 x 107
3.71 x 107
7.76 x 107
2.24 x 107
Mín. 107
Mín. 107
Yeast (Log CFU/mL)
9.33 x 105
1.67 x 106
1.32 x 106
1.00 x 106
Mín. 104
Mín. 104
Titratable acidity (%)
0.83 ± 0.26
0.85 ± 0.16
0.70 ± 0.11
0.75 ± 0.11
0.5 a 1.5
Mín. 0.6
Protein (%)
3.70 ± 0.09
3.67 ± 0.17
3.68 ± 0.07
3.73 ± 0.13
Mín. 2.9
Mín. 2.7
Fat (%)
0.11 ± 0.03
0.11 ± 0.04
0.11 ± 0.04
0.11 ± 0.02
v
< 10
Lactose (%)
3.39 ± 0.42
3.60 ± 0.39
3.57 ± 0.41
3.56 ± 0.47
n.s.
n.s.
pH
4.68 ± 0.28
4.98 ± 0.15
4.69 ± 0.41
4.85 ± 0.22
n.s.
n.s.
RTIQ: Technical Regulation of Identity and Quality of Fermented Milk; V: According to the fat content of the raw material; NS: Not Specified; KLCU-Curitiba; KLDI-Divinópolis; KLSA-Salvador; KLVI-Viçosa.
Table 3: Microbiological and physicochemical features of milk kefir with 48 hours of fermentation (n=4) and current standards.
Sugary water kefir microbiological and physicochemical characteristics
All sugary water kefir beverages reached 107CFU/ml of LAB on 48h, and drop to 106 after 28 days of cold storage (Table 4). Yeast population suffered no significant alterations during storage, maintaining an average level of 106CFU/ml until the end of the experimental period. Titratable acidity has slightly increased during storage (Table 5). The pH reached 4.5 in all sugary water kefir beverages after 24h of fermentation and around 4.0 after storage. Other authors observed a similar drop in pH [3], but lower pH has already found in delayed fermentation time [10].
Microorganism
Sample
Time (Days)
P-Value
1
2
7
28
LAB
(Log CFU/mL)KABH
6.25 ± 1.02a
7.41 ± 0.30a
6.43 ± 0.19a
6.13 ± 0.37a
0.0885
KACU
5.84 ± 0.76a
7.19 ± 0.17a
6.56 ± 0.54a
6.15 ± 1.03a
0.1821
KASA
6.44 ± 0.18c
7.59 ± 0.25a
7.13 ± 0.23ab
6.64 ± 0.15bc
0.0006
KAVI
6.27 ± 0.13ab
7.32 ± 0.41a
7.12 ± 0.70a
6.00 ± 0.10b
0.0112
Yeast
(Log CFU/mL)KABH
6.13 ± 0.49b
6.46 ± 0.17ab
6.70 ± 0.14ab
6.87 ± 0.21a
0.0559
KACU
6.27 ± 0.38a
6.46 ± 0.22a
6.69 ± 0.09a
6.84 ± 0.33a
0.1351
KASA
6.86 ± 0.23a
7.11 ± 0.53a
6.99 ± 0.46a
7.13 ± 0.54a
0.8727
KAVI
5.89 ± 0.80a
6.27 ± 0.43a
6.58 ± 0.55a
6.82 ± 0.52a
0.3143
Different lowercase letters in the lines indicate a significant difference by the Tukey test (P<0.05). KABH-Belo Horizonte; KACU-Curitiba; KASA-Salvador; KAVI-Viçosa.
Table 4: LAB and yeast counts of sugary water kefir produced with grains of different origins, over time (days) (n=3).
Parameter
Sample
Time (Days)
P-Value
1
2
7
28
Titratable acidity (mg lactic acid/100mL)
KABH
0.08 ± 0.03b
0.10 ± 0.02b
0.13 ± 0.03b
0.22 ± 0.03a
0.0025
KACU
0.06 ± 0.02a
0.08 ± 0.01a
0.11 ± 0.01a
0.17 ± 0.03a
0.4921
KASA
0.05 ± 0.01b
0.08 ± 0.02ab
0.13 ± 0.04ab
0.18 ± 0.02a
0.0397
KAVI
0.06 ± 0.03a
0.09 ± 0.03a
0.11 ± 0.02a
0.14 ± 0.02a
0.1666
pH
KABH
4.33 ± 0.50a
4.02 ± 0.19a
3.82 ± 0.08a
3.71 ± 0.21a
0.1141
KACU
4.57 ± 0.22a
4.34 ± 0.12ab
4.12 ± 0.13ab
3.99 ± 0.24b
0.0206
KASA
4.57 ± 0.32a
4.35 ± 0.24a
4.06 ± 0.38a
3.91 ± 0.39a
0.1551
KAVI
4.61 ± 0.55a
4.39 ± 0.45a
4.12 ± 0.34a
4.00 ± 0.40a
0.3848
Different lowercase letters in the lines indicate a significant difference by the Tukey test (P<0.05). KABH-Belo Horizonte; KACU-Curitiba; KASA-Salvador; KAVI-Viçosa.
Table 5: Physicochemical features of water kefir produced with grains of different origins, over time (days) (n=3).
Currently, there are no identity and quality standards defined for sugary water kefir by Brazilian regulatory agencies. According to results of this study and the scientific literature available to this date, standards of 106CFU/mL for lactic acid bacteria and yeast and 0.12mg of lactic acid/mL and pH range of 3.8 to 4.4 could be proposed.
The transcriptionally active microbiome (TAM) of milk and sugary water kefir
In the Bacterial Transcriptionally Active Microbiome (bTAM) of milk kefir (KLSA and KLCU) and sugary water kefir (KASA and KACU), Firmicutes and Proteobacteria phyla comprise more than 99.99% of protein-related reads. However, the relevance of each phylum in milk kefir or sugary water kefir is dissimilar, with Firmicutes and Proteobacteria counting 92% to 97% and 3% to 8%, respectively, of total reads in milk kefir, and 58% to 70% and 30% to 42%, in sugary water kefir (Figure 4A). The families most recurrent were Leuconostocaceae, Lactobacillaceae and Acetobacteraceae (Figure 4B). The bacterial genera most often seen in milk kefir KLSA and KLCU were Leuconostoc (56% and 82%) and Lactobacillus (7% and 39%), and in sugary water kefir KASA and KACU were Oenococcus (26% and 32%), Gluconobacter (15% and 22%) and Lactobacillus (11% and 15%), despite the less accuracy of the description at the genus level in these last kefir samples (Figure 4C). Analyzing bacterial libraries, Leuconostoc mesenteroides (54 and 80%) and Lactobacillus kefiranofaciens (5 and 30%) dominated the milk kefir, while Oenococcus kitaharae (20 and 25%) and Gluconobacter oxydans (15% and 22%) were conspicuous in sugary water kefir, as seen in Figure 4D.
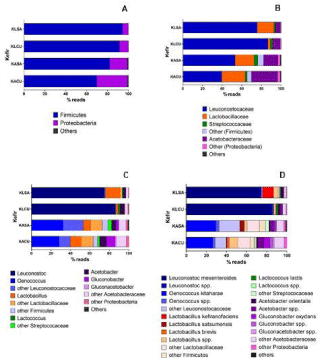
Figure 4: Functional bacteria microbiome in samples of milk kefir (KLSA and KLCU) and sugary water kefir (KASA and KACU) produced with grains of Salvador
and Curitiba, respectively. (A) Fila; (B) Families; (C) Genera; (D) Species.
Regarding Fungal Transcriptionally Active Microbiome (fTAM) of the milk kefir samples, families Pichiaceae, Dipodascaceae, Saccharomycetaceae, and Debaryomycetaceae were the most represented (KLCU, 41%, 38%, 13%, 5%; KLSA, 56%, 22%, 10%, 9%, respectively). In the microbiome of sugary water kefir, Saccharomycetaceae dominated with 92% prevalence in both samples (Figure 5A). The most common genera in the microbiome of milk kefir were Pichia and Yarrowia (KLCU, 36% and 37%; KLSA, 50% and 20%, respectively) (Figure 5B), represented by the species P. fermentans and Y. lipolytica (KLCU, 11% and 35%; KLSA, 25% and 19%, respectively). Analyzing libraries of sugary water kefir, the most found genera were Saccharomyces and Torulaspora (KACU, 61% and 20%; KASA, 65% and 15%, respectively), and the species S. cerevisiae and T. delbrueckii (KACU, 51% and 4%; KASA, 56% and 8%, respectively).
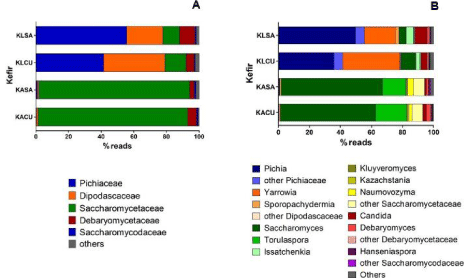
Figure 5: Functional yeast microbiome in samples of milk kefir (KLSA and KLCU) and water kefir (KASA and KACU) produced with grains of Salvador and Curitiba,
respectively. (A) Families; (B) Genera.
The microbial diversity of milk and sugary water kefir has been studied by culture-dependent and culture-independent approaches [2-4,8,10,29,38-40]. This knowledge is essential to understand the community dynamics, metabolite and flavour production beyond such beverages’ rheological characteristics. However, the transcriptomic approach used herein allowed us to elucidate the lactic acid bacteria and yeast species metabolically active in Brazilian kefir and, for instance, effectively responsible for physicochemical characteristics of each beverage.
Conclusion
Brazilian’s milk and sugary water kefir grains possess a matrix associated with its microbiota composed by bacteria and yeasts, but are macroscopically and microscopically distinct, in a similar way to other milk and sugary water kefir grains worldwide. Fermented beverages produced with four Brazilian milk kefir grains from different locations presented similar characteristics that were in accordance with the Codex Alimentarius and Brazilian legislation. This study suggests standards for pH and lactose content values since there is no current official definition for these milk kefir parameters. Fermented beverages produced with four Brazilian water kefir grains from different locations were also similar. This study presented microbiological and physicochemical parameters that can be used as identity standards for this type of fermented beverage since there is no Brazilian legislation that concerns sugary water kefir identity and quality. Differences in the functional microbiome of Brazilian milk and sugary water kefir allow us to conclude that they are different fermented beverages and require establishing specific identity standards for each one by regulatory agencies. Beyond that, knowing the functional microbiome might contribute to the development of appropriate kefir starter cultures.
Acknowledgment
The authors are grateful for financial support of CAPES and FAPEMIG.
References
- Gao X, Li B. Chemical and microbiological characteristics of kefir grains and their fermented dairy products: A review. Cogent Food Agric. 2016; 2: 1272152.
- Leite AMO, Miguel MAL, Peixoto RS, Rosado AS, Silva JT, Paschoalin VMF. Microbiological, technological and therapeutic properties of kefir: a natural probiotic beverage. Braz J Microbiol. 2013; 44: 341-349.
- Magalhães KT, Pereira GVM, Dias DR, Schwan RF. Microbial communities and chemical changes during fermentation of sugary Brazilian kefir. World J Microbiol Biotechnol. 2010; 26: 1241-1250.
- Magalhães KT, Pereira GVM, Campos CR, Dragone G, Schwan RF. Brazilian kefir: structure, microbial communities and chemical composition. Braz J Microbiol. 2011; 42: 693-702.
- Altay F, Karbanglu-Güler F, Dikmen CD, et al. A review on traditional Turkish fermented non-alcoholic beverages: microbiota, fermentation process, and quality characteristics. Int J Food Microbiol. 2013; 167: 44-56.
- Brazil. Ministry of Agriculture Livestock and Supply. Normative Instruction 46. Approves the Technical Regulation of Identity and Quality of Fermented Milks. Diário Oficial da União. Brasília. 2007.
- Codex Alimentarius. Codex standard for fermented milk. Codex Standard 243-2003. Revision, 2018. Roma, 2018.
- Walsh AM, Crispie F, Kilcawley K, O’Sullivan O, O’Sullivan MG, Claesson MJ, et al. Microbialsucession and flavor production in the fermented dairy beverage kefir. Appl Environm Sci. 2017; 1: e00052-e00116.
- Gulitz A, Stadie J, Wenning M, et al. The microbial diversity of water kefir. Int J Food Microbiol. 2011; 151: 284-288.
- Laureys D, De Vuyst L. Kinetics of water kefir fermentation. Appl and Environ Microbiol. 2014; 80:2564-2572.
- Fiorda FA, Melo Pereira GV, Thomaz-Soccol V, Rakshit SK, Pagnoncelli MGB, Vandenberghe LPS, et al. Microbiological, biochemical, and functional aspects of sugary kefir fermentation-A review. Food Microbiol. 2017; 66: 86- 95.
- Abatemarco Junior M, Sandes SHC, Ricci MF, Arantes RM, Nunes AC, Nicoli JR, et al. Protective effect of Lactobacillus diolivorans 1Z, isolated from Brazilian kefir, against Salmonellaenterica serovar Typhimurium in experimental murine models. Front Microbiol. 2018; 9: 2856-2867.
- Farag MA, Jomaa SA, El-Wahed AA, El-Seedi HR. The many faces of kefir fermented dairy products: quality characteristics, flavour chemistry, nutritional value, health benefits, and safety. Nutrients. 2020; 12: 346-368.
- Hamet M, Piermaria JA, Abraham AG. Selection of EPS-producing Lactobacillus strain isolated from kefir grains and rheological characterization of the fermented milk. Food Sci and Technol. 2015; 63: 129-135.
- Paiva IM, Steinberg R, Lula IS, Souza-Fagundes EM, Mendes TO, Bell MJV, et al. Lactobacillus kefiranofaciens and Lactobacillus satsumensis isolated from Brazilian kefir grains produce alpha-glucans that are potentially suitable for food applications. LWT-Food Sci and Technol. 2016; 72: 390-398.
- Leite AMO, Mayo B, Rachid CTCC, Peixoto RS, Silva JT, Paschoalin VMF, et al. Assessment of the microbial diversity of Brazilian kefir grains by PCRDGGE and pyrosequencing analysis. Food Microbiol. 2012; 31: 215-221.
- Miguel MGDCP, Cardoso PG, Lago LDA, Schwan RF. Diversity of bacteria present in milk kefir grains using culture-dependent and culture-independent methods. Food Res Int. 2010; 43: 1523-1528.
- Miguel MGCP, Cardoso PG, Magalhães KT, Schwan RF. Profile of microbial communities present in tibico (sugary kefir) grains from different Brazilian states. World J Microbiol and Biotechnol. 2011; 27: 1875-1884.
- Zanirati DF, Abatemarco Junior M, Sandes SHC, Nicoli JR, Nunes AC, Neumann E. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as a starter or probiotic cultures. Anaer.2015; 32: 70-76.
- Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet.2009; 10: 57-63.
- Botazzi V, Bianchi F. A note on scanning electron microscopy of microorganisms associated with the kefir granule. J Appl Bacteriol. 1980; 48: 265- 268.
- Brazil. Ministry of Agriculture, Livestock and Supply. Normative Instruction 62., Officializes the Official Analytical Methods for Microbiological Analysis for the control of animal products and water. Diário Oficial da União. Brasília. 2003.
- Aronesty E. Comparison of Sequencing Utility Programs. The Open Bioinf J. 2013; 7: 1-8.
- Chen Y, Ye W, Zhang Y, Xu Y. High-speed BLASTN: An accelerated MegaBLAST search tool.Nucl Ac Res. 2015; 43: 7762-7768.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing afull-length transcriptome without a genome from RNASeq data. Nat Biotechnol. 2013; 29: 644-652.
- Tang S, Lomsadze A, Borodovsky M. Identification of protein-coding regions in RNA transcripts. Nucl Ac Res. 2015; 43: e78.
- Fernandes GR, Barbosa DV, Prosdocimi F, Pena IA, Santana-Santos L, Coelho Junior O, et al. A procedure to recruit members to enlarge protein family databases-the building of UECOG (UniRef-Enriched COG Database) as a model. Gen and Mol Res. 2008; 7: 910–924.
- Salvetti E, Harris HMB, Felis GE, O’Toole PW. Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl Environ Microbiol. 2018; 84:e00993-18.
- Garofalo C, Osimani A, Milanovic C, Aquilanti L, De Filippis F, Stelatto G, et al. Bacteria and yeastmicrobiota in milk kefir grains from different italian regions. Food Microbiol. 2015; 49:123-133.
- Garrote GL, Abraham AG, Antoni GL. Chemical and microbiological characterization of kefir grains. J Dairy Res. 2001; 68: 639-652.
- Guzel-Seydim Z, Wyffels JT, Seydim AC, Greene AK. Turkish kefir and kefir grains: microbialenumeration and electron microscopic observation. Int J of Dairy Technol. 2005; 58: 25-29.
- Zhou J, Liu X, Jiang H, Dong M. Analysis of the microflora in Tibetan kefir grains using denaturinggradient gel electrophoresis. Food Microbiol. 2009; 26: 770-775.
- Tomovska J, Gjorgievsky N, Makarijoski B. Examination of pH, titratable acidity and antioxidanteactivity in fermented milk. J Mat Sci Engineer. 2016; 6: 326-333.
- Guzel-Seydim Z, Kok-Tas T, Greene AK. Kefir and koumiss: microbiology and technology. In:Yildiz F, editor. In: Development, and manufacture of yogurt and other functional dairy products. CRCPress. Boca Raton. 2010; 143-163.
- Magra TI, Antoniou KD, Psomas EI. Effect of milk fat, kefir grain inoculum and storage time on theflow properties and microbiological characteristics of kefir. J Text Studies. 2012; 43: 299-308.
- Martins AR, Monteiro RL, Burkert JFM, Burket CAV. Simultaneous enzymatic hydrolysis and lacticfermentation to obtain a yogurt with lactose content. Ciên. Agrotecnol. 2012; 36: 551-559.
- Irigoyen A, Arana I, Castiella M, Torre P, Ibañez FC. Microbiological, physicochemical, and sensory characteristics of kefir during storage. Food Chem. 2005; 90: 613-620.
- Gao W, Zhang L. Comparative analysis of the microbial community composition between Tibetan kefir grains and milks. Food Res Int. 2019; 116:137-144.
- Gulitz A, Stadie J, Ehrman MA, Ludwig W, Vogel RF. Comparative phylobiomic analysis of thebacterial community of water kefir by 16S rRNA gene amplicon sequencing and ARDRA analysis. J Appl Microbiol. 2013; 114: 1082-1091.
- Marsh AJ, O’Sullivan O, Hill C, Ross RP, Cotter PD. Sequencing-Based Analysis of the Bacterialand Fungal Composition of Kefir Grains and Milks from Multiple Sources. PloS One. 2013; 8: e69371.