
Research Article
Austin J Clin Med. 2024; 9(2): 1054.
Transcriptome Analysis of The Potential Mechanisms Regulating Autophagy in Matrine-Treated IMCD3 Cells
Chenghua Yan1#; Feifei Guo1#; Rongliang Wang1; Wendong Kuang2; Ling Niu1; Wu Zaiqiang1; Yongcui Liao1; Guangqiang Ma1,4*; Liang Jin2*
1College of Life Sciences, Jiangxi University of Chinese Medicine, China
2Institute of Microbiology, Jiangxi Academy of Sciences, China
3School of Clinical Medicine, Nanchang Medical College, China
4Key Laboratory of Evaluation of Traditional Chinese Medicine Efficacy (Prevention and Treatment of Brain Diseases with Mental Disorders), Key Laboratory of Depression Animal Model Based on TCM Syndrome, Jiangxi Administration of Traditional Chinese Medicine, Key Laboratory of TCM for Prevention and Treatment of Brain Diseases with Cognitive Dysfunction, China
*Corresponding author: Guangqiang Ma, College of Life Sciences, Jiangxi University of Chinese Medicine, Nanchang 330004, China; Liang Jin, Institute of Microbiology, Jiangxi Academy of Sciences, Nanchang 330029, China. Email: maguangqiang@163.com; jinliang079@163.com
#These authors have been equally contributed to this article.
Received: November 04, 2024; Accepted: November 25, 2024; Published: December 02, 2024
Abstract
Polycystic Kidney Disease (PKD) is a genetic disorder characterized by uncontrolled proliferation of renal cells, with the consequent formation of cysts and loss of renal function. Matrine has the effect of regulating autophagy, and is considered to regulate inflammatory responses and cyst formation. Therefore, in this study we focused on the pathological mechanism of matrine-regulated autophagy in polycystic kidney disease, and identified some autophagy-regulated genes. We also performed transcriptome sequencing of matrine-treated mouse renal epithelial cells (IMCD3). The pathway analysis results showed that signal transduction, including adrenergic signaling in cardiomyocytes, Hippo signaling pathway, and calcium signaling pathway, which are closely related to autophagy, comprises the main pathological changes of IMCD3 cells treated with matrine. These results indicate that exaggerated autophagy participates in the pathological process of polycystic kidney disease, and may provide new insight for further basic research on PKD.
Keywords: Polycystic kidney disease; Matrine; IMCD3 cells; Autophagy
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is a common monogenic inherited kidney disease mainly caused by polycystic kidney disease gene 1 (Pkd1) and polycystic kidney disease gene 2 (Pkd2) mutations. The incidence rate of ADPKD is approximately 1/400-1/1000 [1], and it is more common in adults. This disease is mainly characterized by the appearance of renal tubular epithelial cell cysts, gradual fibrosis of renal parenchyma, and progressive decline of renal function, and eventually leads to renal failure and end-stage renal disease (ESRD). Polycystic Kidney Disease (PKD) is one of the four major causes of end-stage renal disease in China [2]. According to epidemiological statistics, there are approximately 1.5 million patients in China. ADPKD has a high incidence and poor prognosis, thus it has received widespread attention from society and the medical community. Patients are often accompanied by complications such as pain, hematuria, and intracystic infection in clinical practice. Additionally, patients may experience several intracystic infections throughout their lifetime. At present, the commonly used clinical drugs mainly relieve clinical symptoms and delay renal failure by inhibiting cyst expansion and cyst fluid secretion. There is no specific cure drug for PKD. If the disease progresses to end-stage renal disease, then dialysis treatment or kidney transplantation is the main method used. Nearly half of ADPKD patients eventually require renal replacement therapy [3]. Above all, the development of effective drugs to alleviate or treat polycystic kidney disease is a major issue in the medical field. Therefore, research on the pathogenesis of polycystic kidney disease plays an important role in determining effective treatment options. Traditional Chinese medicine is a valuable asset in China and even in the world. Many traditional Chinese medicines or their extracts have shown significant advantages in the treatment of various diseases. Sophora flavescens Ait is a traditional Chinese medicinal herb, which has been used for more than 2,000 years due to its anti-inflammatory, antiviral, antitumor and other functions [4]. The biologically active components of Sophora flavescens mainly include alkaloids and flavonoids, among which matrine has a relatively high content in alkaloids and is the most important active substance [5,6]. Matrine also has antiviral, antitumor, antibacterial and anti-inflammatory effects [7]. ADPKD is also an inflammatory disease characterized by renal cysts. The results of our previous study showed that matrine induces IMCD3 cells autophagy through the MAPK signaling pathway, yet the detailed molecular mechanism requires further study [8]. This study aimed to explore the regulatory mechanism of matrine on autophagy in renal epithelial cells, so as to provide new therapeutic ideas for clinical treatment of patients with polycystic kidney disease.
Recently, With the development of high-throughput sequencing technology, RNA-seq has been widely used in study the pathogenesis of many diseases and their underling regulatory mechanisms [9- 11]. In the present study, IMCD3 cells were treated with different doses of matrine, to perform transcriptomic analysis and reveal the potential effects of matrine on IMCD3 cell autophagy. DEGs in the transcriptome were validated using quantitative reverse transcription PCR (qRT-PCR). These results will improve our understanding of the molecular mechanism(s) underlying the effects of matrine on IMCD3 cells and will be beneficial to developing effective medicine for PKD.
Results
Matrine-Induced IMCD3 Autophagy
We treated cells with different concentrations of matrine for 24 h, and found that, compared with the blank control group, when the concentration of matrine was lower than 0.8 mg/mL, the cell viability did not change significantly, and when the concentration of matrine was 1.6 mg/mL, the cell viability was significantly inhibited, as previously reported8. Therefore, we selected 0, 0.2, 0.4, 0.6 and 0.8 mg/ mL of matrine to treat the cells, so as to detect the effect on autophagy. As shown in Fig. 1, we found that autophagy was significantly enhanced with increasing matrine concentration, but there is little difference between 0.6 and 0.4 concentrations. Based on the above data, to study the specific molecular mechanism of matrine affecting autophagy, 0, 0.2, 0.4 and 0.8 mg/mL of matrine were chosen as the doses in subsequent analyses.
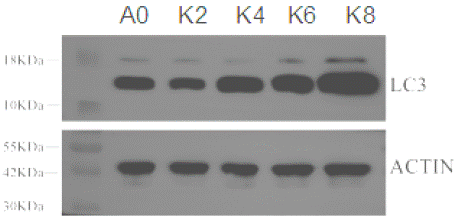
Figure 1: Matrine-induced IMCD3 cell autophagy. IMCD3 cells were treated
with different doses of matrine for 24 h, and LC3 was analyzed by Western
blot(A0: without matrine; K2: 0.2 mg/mL matrine; K4: 0.4 mg/mL matrine; K6:
0.6 mg/mL matrine; K8: 0.8 mg/mL matrine).
Overall Statistical Analyses of the Transcriptome Sequences
Through high-throughput sequencing RNA-Seq of the 16 samples, we obtained 115.79 GB clean data, and the clean data of each sample were above 6.59 GB. Additionally, the Q30 and GC content of the clean reads respectively ranged from 91.87 to 97.4% and 51.37 to 51.96%. The clean reads of each sample were sequenced with the designated reference genome GRCm39 (http://asia.ensembl.org/ Mus_musculus/Info/Index), and the alignment rates ranged from 92.37% to 94.81% (Table 2), thus indicating that the quality of RNAseq was sufficient for the subsequent analysis. After assembly, a total of 31,447 expressed genes were detected, including 30,647 known genes and 800 new genes, and a total of 107,691 expressed transcripts, including 92,262 known transcripts and 15,429 new transcripts.
Table 2: Summary of RNA-seq data.
Significant Impacts of Matrine on Transcriptome Expressions in IMCD3
The principal component analysis of this study showed that the samples of different treatments were clearly separated (Figure 2a), indicating that all of the samples were reproducible, with the exception of A0_1 (Figure S1). Therefore, we removed A0_1 in the following analysis. A heatmap of gene expression is shown in Fig. 2b. The results show that these DEGs can effectively distinguish among the samples, and that the mRNA expressions between the control group and matrine treated group were distinctive. All of the results showed that matrine treatment could significantly affect the gene expression of the IMCD3 cells. In addition, the transcriptome results showed that 46 genes were down-regulated and 46 genes were upregulated as a response to the 0.2 mg/mL matrine treatment; 222 were down-regulated and 229 were up-regulated as a response to the 0.4 mg/mL matrine treatment; and 636 were down-regulated and 640 were up-regulated as a response to the 0.8 mg/mL matrine treatment (Figure 2c).
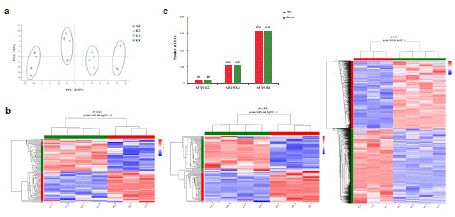
Figure 2: Significant impacts of matrine on transcriptome expressions in
IMCD3. a) Principal Component Analysis (PCA) of different concentration
of matrine treated groups, except for A0_1 (A0: without matrine; K2: 0.2 mg/
mL matrine; K4: 0.4 mg/mL matrine; K8: 0.8 mg/mL matrine). b) Heat map
of differently expressed genes (DEGs) in the matrine-treated samples (K2,
K4, K8) and controls (A0). c) Number of up-regulated genes (red bars) and
down-regulated genes (green bars) following different concentrations of
matrine treatment.
Analysis of the Differently Expressed Genes (DEGs)
In the present study, KEGG enrichment analyses were performed on the DEGs expressed in the different doses of the matrine-treated groups to clarify the relevant biological pathways. According to the pvalue (< 0.05) of KEGG analysis of the up- and down-regulated signal pathways, we identified significant up- and down-regulated signal pathways of the 0.2 mg/mL matrine-treated group compared to the control group (Figure 3a & Figure S2a). The up-regulated DEGs were enriched in the digestive system, including gastric acid secretion and pancreatic secretion. In contrast, the down-regulated genes were enriched in the cholesterol metabolism, influenza A, measles and NOD-like receptor signaling pathway, among which NOD-like receptor signaling has a role in regulating autophagy [12]. As for the 0.4 mg/mL matrine treated group, we found that, compared with the control group, the up-regulated DEGs are enriched in signal transduction, including adrenergic signaling in cardiomyocytes [13,14], Hippo signaling pathway [15,16], Apelin signaling pathway [17] and calcium signaling pathway [18,19], and these are closely related to autophagy. The down-regulated genes were also enriched in signal transduction, including the NOD-like receptor signaling pathway, Il-17 signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and Chemokine and JAK-STAT signaling pathways, among which the NOD-like receptor signaling pathway, IL-17 signaling pathway [20], AGE-RAGE signaling pathway [21] and JAK-STAT signaling pathway [22] regulate autophagy (Figure 3b & Figure S2b). The DEGs between the control group and 0.8 mg/mL matrine-treated group were also analyzed by KEGG categories (Figure 3c & Figure S2c). The up-regulated enriched pathways were mainly related to calcium signaling pathway, Rap1 signaling pathway, apelin signaling pathway and MAPK signaling pathway [23]. In addition to the Rap1 signaling pathway, these pathways have been reported to play important roles in autophagy. The down-regulated genes were enriched in the NOD-like receptor signaling pathway, Il-17 signaling pathway, JAK-STAT signaling pathway and TNF signaling pathway. Furthermore, the Il-17, JAK-STAT and TNF signaling pathways [24] are related to autophagy.
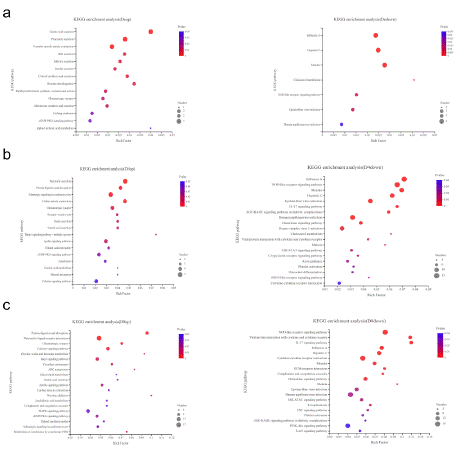
Figure 3: KEGG pathway enrichment analysis of the differently expressed
genes (DEGs). a) Up-regulated pathways (left) and down-regulated
pathways (right) following 0.2 mg/mL matrine treatment. b) Up-regulated
pathways (left) and down-regulated pathways (right) following 0.4 mg/mL
matrine treatment. c) Up-regulated pathways (left) and down-regulated
pathways (right) following 0.8 mg/mL matrine treatment.
From the above results we observed that, compared with the control group, when the concentration of matrine was 0.2 mg/mL, the genes related to external stimuli were mainly altered. With the increasing concentration of matrine, there were significant changes in the genes involved in cell autophagy. This is consistent with our Western blot results, which showed that autophagy was significantly enhanced with increasing matrine concentrations.
Verification of DEGs by qRT-PCR
Autophagy plays an important role in the pathogenesis of PKD. We found that 0.2, 0.4 and 0.8 mg/mL matrine all induced IMCD3 cell autophagy (Figure 1). Next, we compared the DEGs in different doses of the matrine-treated group and selected 78 co-expressed DEGs (Figure 4a). We verified some of these differentially expressed genes obtained from RNA-seq by qRT-PCR (Figure 4b).
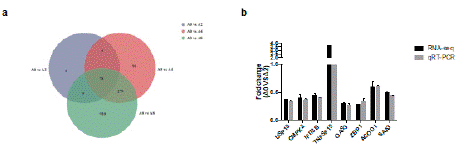
Figure 4: Verification of DEGs by qRT-PCR. a) Venn diagram for the DEGs
in different doses of the matrine-treated samples and controls. b) The relative
mRNA levels of selected DEGs in the matrine treated samples and controls
were examined by qPCR. The mRNA levels by qPCR are presented as the
fold change compared with the A0 sample after normalization against HPRT.
As shown in Fig 4b, for all selected genes, the fold-change values obtained for the matrine-treated sample versus the control sample in the results of the qPCR analysis were consistent with those obtained by RNA-seq.
Verification of DEGs by qRT-PCR
Autophagy plays an important role in the pathogenesis of PKD. We found that 0.2, 0.4 and 0.8 mg/mL matrine all induced IMCD3 cell autophagy (Figure 1). Next, we compared the DEGs in different doses of the matrine-treated group and selected 78 co-expressed DEGs (Figure 4a). We verified some of these differentially expressed genes obtained from RNA-seq by qRT-PCR (Figure 4b). As shown in Fig 4b, for all selected genes, the fold-change values obtained for the matrinetreated sample versus the control sample in the results of the qPCR analysis were consistent with those obtained by RNA-seq.
Discussion
The pathogenesis of polycystic kidney disease is quite complex, and the treatment methods are limited. It has been reported that autophagy is closely related to the pathogenesis of polycystic kidney disease, and is anticipated to be a new method for the treatment of polycystic kidney disease [25,26]. Increasing numbers of studies have focused on the role of renal autophagy in PKD [27-29]. Matrine is a novel autophagy inducer [30-32] that can induce autophagy in IMCD3 cells, but the molecular mechanism of matrine-induced autophagy in IMCD3 cells remains unclear. The goals of this study were to analyze the transcriptomes of the matrine-treated group and the control group, determine the autophagy-related genes that change with the matrine concentration gradient, and reveal the detailed molecular mechanism of matrine-induced autophagy.
Along with the development of sequencing technology and the diversification of sequencing methods, our knowledge of diseases ranges from simple morphology and histopathology to complex molecular pathology, and we now possess a more comprehensive and in-depth understanding of the pathogenesis of diseases. However, at present no studies have completely revealed the pathogenesis of PKD, thus necessitating molecular pathological research on PKD.
In the present study, we treated IMCD3 cells with matrine to induce autophagy, and constructed a new database by means of RNAseq. Through gene differential expression and KEGG analysis and other methods, we found that, with the increase of the matrine dosage, the signaling pathways regulating autophagy gradually underwent significant change. When the concentration of matrine reached 0.4 mg/mL and 0.8 mg/mL, the most highly enriched pathways were the Adrenergic signaling in cardiomyocytes, as well as Hippo, apelin, calcium, MAPK, NOD-like receptor, Il-17, JAK-STAT and TNF signaling pathways, along with AGE-RAGE signaling pathways in diabetic complications (Figure 3b, c). It has been demonstrated that all of these signaling pathways are important in the process of autophagy [33,34]. Alterations in the signals, will drive autophagyrelated diseases such as heart disease [13], neurodegenerative diseases [19] and cancer [15,17]. Furthermore, most of the above signaling pathways are important in the development of PKDs. Recently, the Hippo signaling pathway was found to possibly play a role in Polycystic Kidney Diseases (PKDs) by regulating renal cyst formation [35]. The apelin signaling pathway plays diversified roles in kidney disease, and apelin is a promising therapeutic target for PKD [36]. Calcium signaling participates in kidney cyst formation in ADPKD, and functional loss of either PC1 or PC2 leads to calcium signaling disruption, thus clarifying that the function of calcium may identify new targets for ADPKD treatment [37]. The JAK/STAT signaling pathway regulates cyst growth, and STAT proteins were found to be abnormally activated in the kidneys of PKD [38]. Previous research has identified the TNF superfamily (TNFSF) as a new disease mechanism involved in cystogenesis and cystic growth, and this discovery may lead to a new therapeutic approach in ADPKD [39].
As shown in Figure 1, autophagy was enhanced with the increase of matrine concentration, exhibiting an obvious concentration gradient dependence. Therefore, we selected the genes that altered with the matrine concentration gradient among the 78 genes for further analysis. Among the 78 differentially expressed genes, the expression of 12 genes changed with the concentration of matrine, 10 of which were existing genes in major databases. Four of these 10 genes had been reported to be associated with autophagy, while (TNFSF15, VLDLR, LDLR) are differentially expressed up-regulated genes, and the remaining one (USP18) is a differentially expressed down-regulated gene. TNF superfamily member 15 (TNFSF15) was the most differentially expressed up-regulated gene among all seven genes. Up-regulating TNFSF15 expression leads to increased risk of Inflammatory Bowel Disease (IBD)-induced pyruvate dehydrogenase kinase 1-dependent bacterial uptake, and promotes intracellular bacterial clearance through up-regulation of autophagy in human macrophages [40]. One study reported that the VLDL receptor (VLDLR) facilitates the uptake of triglyceride-derived fatty acids, in turn restraining retinal autophagy. Since fatty acid uptake is reduced in Vldlr-/- mouse tissues, mice are prone to form the neovascular lesions reminiscent of Retinal Angiomatous Proliferation (RAP) [41]. LDLR is a well-known low-density lipoprotein (LDL) receptor closely related to autophagy. Previous research has found that cholesterol accumulation in lysosomes via up-regulated LDL receptor (LDLR) expression, thus significantly reducing cell autophagy by inhibiting of LC3 maturation, p62 degradation and autophagosome formation [42- 44]. Ubiquitin specific peptidase 18 (USP18) is a positive regulator of autophagy and autophagic flux, due to its catalytic activity. Paclitaxel has been reported to down-regulate USP18 expression and inhibit autophagy, thereby overcoming paclitaxel resistance in breast cancer45. Finally, DeISGylation of BECN1 by USP18 promotes the activity of class III PtdIns 3-kinase and autophagy stimulated by type I IFN46. Overall, the differentially expressed up-regulated genes VLDLR and LDLR inhibited autophagy, while the differentially expressed down-regulated gene USP18 promoted autophagy. These results were inconsistent with our experimental results, wherein matrine concentration-dependently induced autophagy. Only the observation that the differentially expressed up-regulated gene TNFSF15 promotes autophagy was consistent with our matrine treatment results. Therefore, we speculate that matrine may promote autophagy by regulating the expression of TNFSF15. In the future, we will further explore the role of TNFSF15 in matrine-induced autophagy.
In conclusion, we identified differentially expressed genes in the untreated and matrine-treated groups. We also identified 12 genes that changed with the concentration gradient of matrine, and observed that matrine may have regulated the TNFSF15 gene, which is closely related to autophagy. The results of this study may provide new insights into understanding the molecular mechanism of autophagy in polycystic kidney disease.
Materials and Methods
Cell Lines and Culture
IMCD3 cell (Shanghai Shunran Biotechnology Co., Ltd.) was inoculated in a high-glucose medium (Solebo, 12100) containing 10% fetal bovine serum (GEMINI, 900-108) and 1% penicillin and streptomycin at 37°C in a 5% CO2 incubator.
Sample Preparation
The IMCD3 cells treated with different doses of matrine were collected after 24 h, and the cells were then immediately washed in 0.8% Diethylpyrocarbonate (DEPC)-treated physiologic saline solution to remove the attached leaf pieces, then frozen in liquid nitrogen. The total RNA was extracted for transcriptomic analyses.
RNA Isolation, cDNA Library Preparation and RNA-seq
All of the procedures of RNA-seq were conducted by Majorbio Co. (Shanghai, China). The total RNA was isolated from the IMCD3 cells using Trizol reagent (Life Technologies) following the manufacturer’s protocols. The RNA integrity and concentrations were examined with an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and Nanodrop2000. The mRNA was enriched with Oligo(dT) magnetic beads (Epicentre, USA), and fragmented into short fragments using a fragmentation buffer. These short fragments were reverse transcribed into cDNAs with random primers to create cDNA libraries. The obtained cDNA libraries were sequenced on the Illumina Novaseq 6000 platform.
Assembly and Functional Annotation
The raw data were filtered with the FASTQ_Quality_Filter tool from the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_ toolkit/, Version 0.0.14). Next, Trinity software (https://github.com/ trinityrnaseq/trinityrnaseq, v2.8.5) was used to assemble all sample clean data from scratch. TransRate (http://hibberdlab.com/transrate/, v1.0.3) 30 and BUSCO (Benchmarking Universal Single-Copy Orthologs, http://busco.ezlab.org, v3.0.2) were used to optimize and evaluate the assembly. All assembled transcripts were compared with six major databases (NCBI_NR, NCBI non-redundant protein library; Swiss-Prot; PFAM; COG, clusters of orthologous groups of proteins; GO, gene ontology; and KEGG, Kyoto Encyclopedia of Genes and Genomes) to obtain the annotated information for each transcript.
Analysis of Differentially Expressed Genes
The transcriptome data were analyzed using the free online tools of the Majorbio Cloud Platform (http://www.majorbio.com). The FPKM (fragments per kb per million reads) method [25] was used to calculate the expression levels of the unigenes, thus eliminating the effects of different gene lengths and sequencing levels. The differentially expressed genes (DEGs) were analyzed by DESeq2 (http://bioconductor. org/packages/stats/bioc/DESeq2/, v1.24.0) as described by Love et al [47]. The significance threshold for the differential expression was padjust < 0.05 and |log2FC|> = 1. Next, all DEGs were subjected to enrichment analysis for the GO function and KEGG pathway, which were considered significantly enriched at P value_uncorrected < 0.05.
Validation of Candidate Genes by qRT-PCR
Next, we used qRT-PCR to verify the DEGs recognized by transcriptome. The total RNA was extracted from the transcriptomic analyzed samples using Trizol reagent (Life Technologies), then treated with DNase I (Fermentas, Glen Burnie, MD, USA). We reverse transcribed 1 μg of the total RNA per sample into complementary DNA (cDNA) using a PrimeScript RT Reagent Kit (Takara). Realtime PCR was performed using Talent qPCR Pre-Mix SYBR Green (Tiangen, China) on a QuantStudioTM6 Flex Real-Time PCR System (Applied BiosystemsTM). The expression levels of each gene were calculated using the 2-ΔΔCt method, and HPRT served as the reference gene. All of the primers used are listed in Table 1. All results are representative of two to three independent biological experiments. The data were analyzed with GraphPad Prism 6.
Table 1: qRT-PCR primers.
Data Availability Statement
The datasets generated during the current study are available in online repository. The names of the repository and accession number can be found below: NCBI under accession PRJNA883545 for data of RNA-seq.
Author Statements
Author Contributions
CY analyzed the data, generated figures and wrote the paper. WK, RW, LN and FG performed the experiments. GM designed the experiments and analyzed the data. LJ was responsible for revising the manuscript. All Authors have read and approved the final version of the manuscript.
# Chenghua Yan and Feifei Guo have contributed equally to this work.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81960808), Department of Education Technology Project of Jiangxi (GJJ211248), Doctoral Research Startup Fund of Jiangxi University of Chinese Medicine(2020BSZR012) and Jiangxi University of Chinese Medicine Science and Technology Innovation Team Development Program (CXTD22013).
Competing Interests
The authors declare no competing interests.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81960808), Doctoral Research Start-up Fund of Jiangxi University of Chinese Medicine(2020BSZR012) and Jiangxi University of Chinese Medicine Science and Technology Innovation Team Development Program (CXTD22013).
References
- Bergmann C, Guay-Woodford LM, Harris PC, Horie S, Peters DJ, Torres VE, et al. Polycystic kidney disease. Nat Rev Dis Primers. 2018; 4: 50.
- Zhou C, Mei C, Xue C. Preimplantation Genetic Diagnosis of Autosomal Dominant Polycystic Kidney Disease Applied in China. Am J Kidney Dis. 2018; 72: 767.
- Takiar V, Nishio S, Seo-Mayer P, King JD, Li H, Zhang L, et al. Activating AMP-activated protein kinase (AMPK) slows renal cystogenesis. Proc Natl Acad Sci USA. 2011; 108: 2462-2467.
- Li JJ, Zhang X, Shen XC, Long Q, Xu CY, Tan CJ, et al. Phytochemistry and biological properties of isoprenoid flavonoids from Sophora flavescens Ait. Fitoterapia. 2020; 143: 104556.
- Unger E, Bohm KJ, Vater W. Factors regulating microtubule structure--a minireview. Acta Histochem. 1986; 33: 85-94.
- You L, Yang C, Du Y, Liu Y, Chen G, Sai N, et al. Matrine Exerts Hepatotoxic Effects via the ROS-Dependent Mitochondrial Apoptosis Pathway and Inhibition of Nrf2-Mediated Antioxidant Response. Oxid Med Cell Longev. 2019; 2019: 1045345.
- Zhang H, Chen L, Sun X, Yang Q, Wan L, Guo C,. Matrine: A Promising Natural Product with Various Pharmacological Activities. Front Pharmacol. 2020; 11: 588.
- Guangqiang Ma LN, Yingying Song, Hongjiao Wan, Liang Jin, Chenghua Yan. Study on mechanism of matrine induces autophagy of IMCD3 cells through MAPK/mTOR signaling pathway. Chinese Journal of Immunology (Chinese Journal). 2022; 38: 282-287.
- Zhao M, Jiang J, Zhao M, Chang C, Wu H, Lu Q,. The Application of Single- Cell RNA Sequencing in Studies of Autoimmune Diseases: a Comprehensive Review. Clin Rev Allergy Immunol. 2021; 60: 68-86.
- Gonzalez-Silva L, Quevedo L, Varela I. Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer. 2020; 6: 13-19.
- Garcia S, Der E, Putterman C. Single Cell RNA Sequencing in Human Disease: Renal, Pancreatic, and Viral Diseases. Adv Exp Med Biol. 2020; 1255: 195-202.
- Pei G, Dorhoi A. NOD-Like Receptors: Guards of Cellular Homeostasis Perturbation during Infection. Int J Mol Sci. 2021; 22: 6714.
- Kumari R, Ray AG, Mukherjee D, Chander V, Kar D, Kumar US, et al. Downregulation of PTEN Promotes Autophagy via Concurrent Reduction in Apoptosis in Cardiac Hypertrophy in PPAR alpha(-/-) Mice. Front Cardiovasc Med. 2022; 9: 798639.
- Deng J, Jiang P, Yang T, Huang M, Xie J, Luo C, et al. beta2adrenergic receptor signaling promotes neuroblastoma cell proliferation by activating autophagy. Oncol Rep. 2019; 42: 1295-1306.
- Wang D, He J, Huang B, Liu S, Zhu H, Xu T. Emerging role of the Hippo pathway in autophagy. Cell Death Dis. 2020; 11: 880.
- Tang F, Christofori G. The cross-talk between the Hippo signaling pathway and autophagy: implications on physiology and cancer. Cell Cycle. 2020; 19: 2563-2572.
- Lv D, Luo X, Chen Z, Liu J, Liu M, Li Y, et al. Apelin/APJ signaling activates autophagy to promote human lung adenocarcinoma cell migration. Life Sci. 2021; 281: 119763.
- Bootman MD, Chehab T, Bultynck G, Parys JB, Rietdorf K. The regulation of autophagy by calcium signals: Do we have a consensus? Cell Calcium. 2018; 70: 32-46.
- Sukumaran P, De Conceicao VN, Sun Y, Ahamad N, Saraiva LR, Selvaraj S, et al. Calcium Signaling Regulates Autophagy and Apoptosis. Cells. 2021; 10: 2125.
- Zhao Y, Li Y, Wang J, Manthari RK, Wang J. Fluoride induces apoptosis and autophagy through the IL-17 signaling pathway in mice hepatocytes. Arch Toxicol. 2018; 92: 3277-3289.
- Shi M, Yang S, Zhu X, Sun D, Sun D, Jiang X, et al. The RAGE/STAT5/ autophagy axis regulates senescence in mesangial cells. Cell Signal. 2019; 62: 109334.
- Xu J, Zhang J, Mao QF, Wu J, Wang Y. The Interaction Between Autophagy and JAK/STAT3 Signaling Pathway in Tumors. Front Genet. 2022; 13: 880359.
- Zhou YY, Li Y, Jiang WQ, Zhou LF. MAPK/JNK signalling: a potential autophagy regulation pathway. Biosci Rep. 2015; 35: e00199.
- Yuan Y, Ding D, Zhang N, Xia Z, Wang J, Yang H, et al. TNF-alpha induces autophagy through ERK1/2 pathway to regulate apoptosis in neonatal necrotizing enterocolitis model cells IEC-6. Cell Cycle. 2018; 17: 1390-1402.
- Tang C, Livingston MJ, Liu Z, Dong Z. Autophagy in kidney homeostasis and disease. Nat Rev Nephrol. 2020; 16: 489-508.
- Aguilar A. Polycystic kidney disease: Autophagy boost to treat ADPKD?. Nat Rev Nephrol. 2017; 13: 134.
- Lee EJ, Ko JY, Oh S, Jun J, Mun H, Lim CJ, et al. Autophagy induction promotes renal cyst growth in polycystic kidney disease. E Bio Medicine. 2020; 60: 102986.
- Dong K, Zhang C, Tian X, Coman D, Hyder F, Ma M, et al. Renal plasticity revealed through reversal of polycystic kidney disease in mice. Nat Genet. 2021; 53: 1649-1663.
- Liu G, Kang X, Guo P, Shang Y, Du R, et al. miR-25-3p promotes proliferation and inhibits autophagy of renal cells in polycystic kidney mice by regulating ATG14-Beclin 1. Ren Fail. 2020; 42: 333-342.
- Cho YR, Lee JH, Kim JH, Lee SY, Yoo S, Jung MK, et al. Matrine suppresses KRAS-driven pancreatic cancer growth by inhibiting autophagy-mediated energy metabolism. Mol Oncol. 2018; 12: 1203-1215.
- Wu J, Hu G, Dong Y, Ma R, Yu Z, Jiang S, et al. Matrine induces Akt/mTOR signalling inhibition-mediated autophagy and apoptosis in acute myeloid leukaemia cells. J Cell Mol Med. 2017; 21: 1171-1181.
- Du J, Li J, Song D, Li Q, Li L, Li B, et al. Matrine exerts antibreast cancer activity by mediating apoptosis and protective autophagy via the AKT/mTOR pathway in MCF7 cells. Mol Med Rep. 2020; 22: 3659-3666.
- Xu Z, Han X, Ou D, Liu T, Li Z, Jiang G, et al. Targeting PI3K/AKT/mTORmediated autophagy for tumor therapy. Appl Microbiol Biotechnol. 2020; 104: 575-587.
- Kma L, Baruah TJ. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol Appl Biochem. 2022; 69: 248-264.
- Ren Z, Zhang Z, Liu TM, Ge W. Novel zebrafish polycystic kidney disease models reveal functions of the Hippo pathway in renal cystogenesis. Dis Model Mech. 2021; 14: dmm049027.
- Chapman FA, Nyimanu D, Maguire JJ, Davenport AP, Newby DE, Dhaun N. The therapeutic potential of apelin in kidney disease. Nat Rev Nephrol. 2021; 17: 840-853.
- Mangolini A, de Stephanis L, Aguiari G. Role of calcium in polycystic kidney disease: From signaling to pathology. World J Nephrol. 2016; 5: 76-83.
- Strubl S, Torres JA, Spindt AK, Pellegrini H, Liebau MC, Weimbs T. STAT signaling in polycystic kidney disease. Cell Signal. 2020; 72: 109639.
- Cordido A, Nunez-Gonzalez L, Matinez-Moreno JM, Lamas-Gonzalez O, Rodriguez-Osorio L, Perez-Gomez MV, et al. TWEAK Signaling Pathway Blockade Slows Cyst Growth and Disease Progression in Autosomal Dominant Polycystic Kidney Disease. J Am Soc Nephrol. 2021; 32: 1912- 1932.
- Sun R, Hedl M, Abraham C. TNFSF15 Promotes Antimicrobial Pathways in Human Macrophages and These Are Modulated by TNFSF15 Disease-Risk Variants. Cell Mol Gastroenterol Hepatol. 2021; 11: 249-272.
- Heckel E, Cagnone G, Agnihotri T, Cakir B, Das A, Kim JS, et al. Triglyceridederived fatty acids reduce autophagy in a model of retinal angiomatous proliferation. JCI Insight. 2022; 7: e154174.
- Lee SJ, Choi YJ, Kim HI, Moon HE, Paek SH, Kim TY, et al. Platycodin D inhibits autophagy and increases glioblastoma cell death via LDLR upregulation. Mol Oncol. 2022; 16: 250-268.
- Lu J, Chen PP, Zhang JX, Li XQ, Wang GH, Yuan BY, et al. GPR43 activationmediated lipotoxicity contributes to podocyte injury in diabetic nephropathy by modulating the ERK/EGR1 pathway. Int J Biol Sci. 2022; 18: 96-111.
- Zhu L, Wu G, Yang X, Jia X, Li J, Bai X, et al. Low-density lipoprotein mimics insulin action on autophagy and glucose uptake in endothelial cells. Sci Rep. 2019; 9: 3020.
- Ge X, Zhang D, Song S, Mi Y, Shen Y, Jiang Q, et al. USP18 reduces paclitaxol sensitivity of triple-negative breast cancer via autophagy. Biochem Biophys Res Commun. 2022; 599: 120-126.
- Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, et al. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015; 11: 617-628.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550.