
Research Article
Austin J Biotechnol Bioeng. 2021; 8(1): 1110.
Antagonistic Activity of Indigenous Rhizobacteria through Biosynthesis of Indole-3-Acetic Acid (IAA), Hydrogen Cyanide (HCN), and Siderophores
Aye Khaing1,2, Theint Theint Win1,2, Kay Thi Oo1,2 and Pengcheng Fu1*
¹State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, Haikou, Hainan Province, China
²Department of Biotechnology Research, Ministry of Education, Kyaukse, 05151, Myanmar
*Corresponding author: Pengcheng Fu, State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, 58 Renmin Avenue, Haikou, Hainan Province 570228, China
Received: May 31, 2021; Accepted: June 29, 2021; Published: July 06, 2021
Abstract
This study aimed to isolate indigenous Antagonistic Bacteria (AB) against common soil-borne phytopathogens, including Rhizoctonia solani, Phythium sp., Fusarium oxysporum. Biosynthesis of Indole-3-Acetic Acid (IAA), generation of Hydrogen Cyanide (HCN), and siderophores production were assessed for their involvement in the antagonistic activities. Rhizospheric soil of bean roots, sunflowers, wheat, rice, and humic and semi flooded soils were used to isolate twenty-one bacterial strains for phytopathogenic antagonism. It was found that nine isolates have potential antagonistic activity against three common soilborne pathogens. Antifungal productivity for IAA, HCN, and siderophores was screened on the isolates while nine ABs were identified. To choose statistically significant isolate for the formulation, the principal component analysis was performed with five variables (IAA production, siderophores production index, antagonistic activities against three phytopathogens). Among the nine isolates, the isolate Pseudomonas alicaligenes shew a positive correlation with all variables. In particular, the strain demonstrated to be an antagonistic strain against the fungal pathogens and a strong producer of siderophores, HCN and IAA, We prepared the biocontrol agents with rice flour, glutinous rice flour, Monosodium Glutamate and Chitosan that were found to maintain the up to three months for convenience of field applications.
Keywords: Antagonistic bacteria; Phytopathogen; Siderophore; Indole-3- acetic acid; Hydrogen cyanide; Soil-borne
Introduction
Agriculture is a critical sector in national economy for developing countries like Myanmar. In order to enhance crop productivity, chemical fertilizers and pesticides are applied to achieve higher yields for the income of farmers, although it contributes to the destruction of microbial species and ecosystems. It is also deposited pollutants in the environment, causing contamination, bioaccumulation, and biomagnifications in the farmlands. To be worse, fertilizer abuse can not only impact soil health, but also promote the growth of soil-borne pathogens which causes heavy loss in crop harvest [1-4]. The immense use of chemical fertilizers in agriculture has even become threats to consumer health [5]. Accordingly, the attention in environmentally sustainable farming techniques and organic farming has increased [6].
Capable of suppressing plant disease, plant growth Rhizobacteria are used in agriculture as a substitute to agrochemicals to increase crop quality and yield through different mechanisms, i.e. Production of siderophores, HCN, antibiotics, antifungal enzymes, vying for colonisation sites, nutrients, and induction of pervasive stress tolerance [6-9]. The Native microbial compositions are classes of innate microbial consortia, which inhabit in soil. They are capable of biodegradation, nitrogen fixation, and enhancement for soil fertility. In particular, Native microbial compositions play an important role in protection of crops from invasion of soil-borne plant pathogens by producing bacteriocins and other inhibitory substances. They contend with the plant pathogens for vital nutrients and host cell receptors, which will block the pathways for the soil-borne pathogens in the rhizosphere to enter higher plants through their roots [10,11].
Among the bacterial metaboilites, siderophores are a group of small biomolecules with high Fe3+ scavenging ability [12] which are capable for chelation and transportion of this element into bacterial cells. Siderophores have been effectively used as a biocontrol agent against several soil-borne phytopathogens. It would significantly lessen the amount of Fe accessible to microflora of the rhizosphere and hinder fungal growth [13]. In addition, down-regulation of IAA as signaling is attributed to plant defence mechanisms against a wide range of phytopathogenic bacteria [14,15]. Hydrogen Cyanide (HCN) is a broad-spectrum antimicrobial compound that is associated with the biological control of root diseases [16]. It has been implicated in the inhibition of various root diseases in tobacco, cucumber and tomato [17]. The importance of HCN in biocontrol has been studied by several researchers in the topics such as antifungal activity [18-20], nematocidal activity [21,22].
There are two objectives for this study: the first one at the mocribial level to isolate and screen indigenous antagonistic bacteria from different rhizospheric soils. The second one was at metabolic level to screen the antagonistic related compound biosynthesis (IAA, HCN, siderophores) and to formulate the most efficient preserving agents for selected antagonist.
Materials and Methods
Soil samples collection and isolation of bacterial strains
Rhizospheric soil samples have been collected from various crop lands, such as wheat, corn, bean root and sunflower lands, and flooded/humic soilx from various locations around the Kyaukse district of the Mandalay region of Myanmar. The pH of the soil samples was between 7.5 and 8.8, and the temperature between 28 and 35°C. Microbial species have been isolated by a serial dilution and steak plate method. One gram of dried soil was weighed and applied in a sterile test tube to 9 ml of double-distilled water (dd H2O) and shaken well using a vortex mixer; this stock solution was then diluted serially up to 10-7, and 0.1 mL of diluted sample dilution was inoculated on LB (Luria Bertani) media surface and incubated for 2 days at 35°C [23]. Following the Hans Gram staining method, the pure colonies were colonially and microscopically examined and preserved as glycerol stock for further uses [24].
Pathogenic fungal strains
The soil-borne pathogens Rhizoctonia solani, Phythium sp., Fusarium oxysporum were used in these research projects. The first two were collected from the Biotechnology Research Department in Myanmar and the second one supported by the state key laboratory of Marine Resource Utilization in South China Sea, Hainan University, China.
Screening of antagonistic activity
The antagonistic activities of all twenty-one isolates were tested in dual culture against common soil-borne plant pathogenic fungi such as Fusarium oxysporum, Phythium sp., and Rhizotonia solani. The pathogens were inoculated in the middle of the PDA-containing Petri plate, and bacteria were streaked 3 cm away from either side of the pathogen and incubated for 7 days at 28°C. The experiments were performed in triplicate and used a petri plate inoculated with a pathogen alone in the absence of an antagonist as a control. The inhibitory effect of control growth (absence of antagonists) was monitored by determining the radial growth of fungal mycelium on each plate and using the formula:
Inhibition of mycelia growth (%) = X-Y/ X × 100 [3].
Molecular identification of antagonistic isolates
The genomic DNA of nine antagonistic isolates was extracted using the TIANamp Bacteria DNA Kit of TIANGEN Company as instructed by the manufacturer. Genomic DNA was amplified by the amplification of universal primers; 27F (5-AGTTTGATC TTGGCTCAG-3) and 1492 R (5-GTTACCTTGTTACGACTT-3). The resulting amplicons were sequenced by 3730 XL DNA analyzers (Applied Biosystems, USA). The NCBI BLAST database (https://blast. ncbi.nlm.nih.gov/Blast.cgi?PAGE= Nucleotides) was used to perform sequence recognition.
Siderophore activity by plate screening method
Nine isolated strains were grown on modified azurol-S chromium agar plates as defined by [25]. In this experiment, calcium sulfate (60.5 mg) was dissolved in 50 mL of deionized water and the resulting solution was added to Fe (III) solution (10mL). This solution was slowly combined with Hexadecyltrimethylammonium Bromide (HDTMA) (72.9 mg) dissolved in 40 mL of water during stirring. The dark-blue solution that resulted was autoclaved, cooled to 50/60°C, and mixed with 15 g L-1 agar containing 900 mL of sterile MM9 [26]. The bacterial strain was inoculated and incubated in the dark after solidification of the medium on Petri plates (28°C for 5 days). The emergance of the yellow-orange area from the dark blue around the colonies was positive. The zone index for siderophore activity has been determined using the formula;
Index = (colony + zone) diameter/colony diameter.
Characterization of siderophores
Fiss Minimal Medium (FMM) was prepared as given by R. Pal and K. Gokarn (2010), to test the production activity of bacterial siderophores; 5 g of KH2PO4 was dissolved in 924 ml ddH2O, adjusted the pH at 6.8 with 6 M NaOH solution, 10 ml 0.001 gL-1 MnSO4 solution, 10 ml 0.4 gL-1 MgSO4 solution, 10 ml 0.005 gL-1 solution ZnCl2. Sterilized, the medium cooled (60°C). The cooled medium was added with 10 ml of 50 gL-1 sterile glucose solution, and mixed with the medium L-asparagine solution (5 gL-1 asparagine dissolved in 30 ml ddH2O). The isolates were incubated for 2 days in a water bath at 27°C. After incubation, the samples were centrifuged at a rate of 10000 rpm at 21°C for 10 minutes. Further experiments were donducted to identify specific siderophores [27]. The siderophore was classified as catechol or hydroxamate using the following procedure:
Catecolate type of siderophore was observed by using the Arnow test [28], which selectively detects the simple catechol-type siderophores. In this study, the catechol interacts in the presence of nitric acid by establishing a yellow color and becomes red in excess of sodium.
Hydroxamate type of siderophore was screened by ferric perchlorate (Atkin) assay in which Atkin reagent (0.0771 g of FeClO4 in 100 ml of water with 1.43 ml of HClO4) was used. In this experiment, 2.5 ml of Atkin reagent was added to 0.5 ml of supernatant after centrifugation and incubated at room temperature for 5 minutes [29]. Orange colour development indicates a positive reaction to hydroxate siderophore, while pale yellow is a negative reaction.
Screening of Hydrogen Cyanide (HCN) production by antagonestic isolates
Feigl-Anger paper method, based on Reddy et al. (2016), was used for the screening of HCN with isolates. The reaction was prepared by soaking a filter paper in a chloroform solution mixture containing equal volumes of 1% (w/v) copper ethylacetoacetate and 1% (w/v) 4, 4’-methylenebis (Sigma-Aldrich). In a 24-well microtiter plates which contains about 500 μl of prepared metabolite extract (prepared in section 2.2.2) was covered with the Fiegl-Anger paper and sealed airtight with a cover using binding clips. The plate was chilled at -20°C for 1 h and defrosted at room temperature for 30 min to develop clear blue spots on each reaction which is the positive reaction of HCN.
Screening for Indole Acetic Acid (IAA) production by antagonistic isolates
In order to assess IAA, the key auxin associated with physiological processes in many plants, pure nine antagonistic isolates were grown aseptically in LB broth comprising tryptophan (100 mgL-1). These inoculated media were then incubated at 180-200 rpm at 30°C for 48 hours on the rotary shaker. One ml of filtrate was mixed with 2 ml of Salkowski’s reagent (20 ml of 35 per cent perchloric acid: 2 ml of 0.5 N FeCl3) and incubated for 30-120 min in a room temperature under dark conditions. The Pink colour showed IAA production and an optical density of 530 nm was measured using the UV-VIS spectrophotometer [30]. The quantity of generated IAA was linked to a prepared standard curve.
Carrier formulation and shelf life of the selected isolate
Bio-formulations of selected isolates have been preserved by combining broth culture with previously sterilized (rice flour, monosodium glutamate, chitosan and glutinous rice flour) through a variety of combinations. These tablets have been made and kept at room temperature. Regular intervals 30 days to 3 months from the date of mixing were used to measure the shelf life of the formulations. A serial dilution of the agar plate method was used to count the colony-forming unit (cfu). Twelve types of carrier systems have been planned as follows:
T-1: 5 g of rice flour and 0.125 g of Monosodium Glutamate (MSG)
T-2: 5g of rice flour, 0.125g of MSG and 0.02 g of chitosan
T-3: 5g of glutinous rice flour and 0.125 g of MSG
T-4: 5 g of glutinous rice flour, 0.125 g of MSG and 0.02 g of Chitosan
T-5: 2.5 g each of rice flour and glutinous rice flour and 0.125 g of MSG
T-6: 2.5 g each of rice flour and glutinous rice flour, 0.125 g of MSG and 0.02 g of Chitosan
T-7: 5 g of rice flour
T-8: 5 g of glutinous rice flour
T-9: 2.5 g each of rice flour and glutinous rice flour
T-10: 2.5 g each of rice flour and skimmed milk
T-11: 2.5 g each of glutinous rice flour and glutinous rice flour
T-12: 5g of skimmed milk
Statistical analysis
All studies have been performed in at least three biological replications. Each sample was typically calculated in three technical repeats. Minitab 17 statistical software was used to interpret quantitative data (Minitab Ltd., Pennsylvania State University). Data were Analyzed using One-Way Variance Analysis (ANOVA). All results were viewed as means (SD), which suggests that not sharing the same letter is statistically different (p0.05). The Prcomp function in the R software stats package was used to perform multivariate Principal Component Analysis (PCA) (R Core Team, 2013).
Results and Discussion
Isolation, screening of antagonistic activity and identification of antagonistic isolates
The present study is focused on the exploration of possible biofertilizer strains from rhizospheric soil samples from the Kyaukse region of Myanmar. 21 bacterial strains have been isolated from 10 separate sample sites. Isolation was carried out based on various cultural morphologies to screen antagonistic activity using a dual culture system.
Dual plate inhibition experiments were performed using PDA, and significant differences in mean growth diameter were determined using the Turkey test (Table 1). Among the 21 strains analyzed, 9 of the strains listed in Table 1 showed an inhibitory effect on the soil-borne plant phytopathogens Rhizoctonia solani, Phythium sp., Fusarium oxysporum. The isolates were coded as BR1, BR2 (from Bean root rhizosphere), WM, WM2 (from wheat rhizosphere), FS1 and FS2 (from floated soil), HS1, HS2 (from humic soil) and SF (from sunflower rhizosphere). In the present analysis, growth inhibition against Fusarium oxysporum, isolates of FS3 possessed the highest activity (72.3±1.1 per cent). For Phythium sp., BR1 and FS3 had the largest reduction (51.6±1.7 and 56.7±1.3 per cent). WM2 had the highest inhibition against Rhizoctonia solani (66.7±1.2). The growth inhibition tests of WM isolates against three phytopathogens had yielded similar results. It is seen that the maximum growth inhibition of 62.7±0.7 per cent of Fusarium oxysporum, followed by 56.13±1.3 for Phythium sp. and 30.13±0.5 per cent for Rhizoctonia solani. Bacterial antagonism is a dynamic, well-known and well-established mechanism [31]. Antagonistic exploration has shown that the influent growth of bacteria had hindered the growth of fungal pathogens [6]. After additional 10-day incubation, pathogenic mycelia were no longer cover the surface of the inhibition ring, suggesting that the antagonism was very high [32]. Antagonistic testing revealed that confluent bacterial growth inhibited the development of fungal phytopathogens [31,6].
Rhizoctonia solani
Pythium sp.
Fusarium Oxysporum
BR1
38.3±0.6ab
51.6±1.7b
63.9±0.6bc
BR2
45.4±2.7ab
12.5±1.1g
58.3±0.9d
FS2
34.3±0.9ab
28.5±0.7f
67.1±0.9b
FS3
44.9±0.4ab
56.4±0.9a
72.3±1.1a
HS1
33.8±0.7ab
45. 3±0.8c
62.9±1.6c
HS2
32.3±1.8ab
45.1±0.6c
73.5±1.5a
SF
27.9±0.9b
38. 6±0.9d
61.1±0.9cd
WM
30.5±0.4b
56.6±1.3a
62.7±0.7c
WM2
66.7±1.2a
34.3±0.9e
63.1±1.3c
Table 1: The Phytopathogen inhibition activity of Isolates.
Among the 21 isolates, nine isolates with antagonestics activity were identified through cultural, microscopic and 16S rRNA gene sequences. The microscopic morphology of nine antagonistic isolates was shown in Figure 1. Each strain’s 16S rRNA was sequenced and classified using BLAST on the NCBI database (www.ncbi.nlm.nih. gov/BLAST). As shown in Table 2, the 16S rRNA matches 6 different Pseudomonas spp., Leclercia sp., Citrobacter sp., Enterobacter sp.. Also, Table 2 summarized the isolation source, cultural, microscopic, and their accession number [23], we thus selected Pseudomonas sp. as the biocontrol agent to overcame the existing tailback for their commercial use.
Figure 1: Microscopic Morphology of antagonistic isolates (a) HS1 (b) HS2 (c) BR1 (d) BR2 (e) FS2 (f) FS3 (g) SF (h) WM (i) WM2.
No.
Isolates
Strains
Accession No.
Isolation source
Colonial morphology
Microscopic morphology (shape)
1
BR1
Pseudomonas plecoglossicida
MT349504
Bean root
White, mucoid
G-ive, rod
2
BR2
Pseudomonas furukawaii
MT355447
Bean root
Brown, rhizoid
G-ive, rod
3
FS3
Pseudomonas alcaligenes
MT355448
Flooded soil
Light green
G-ive, rod
4
SF
Pseudomonas oleovarans
MT355450
Sunflower
Brown, rhizoid
G-ive, rod
5
WM
Pseudomonas alicaligenes
MT355451
Wheat plant
Deep Brown, rhizoid
G+ive, rod
6
WM2
Pseudomonas mendocina
MT355493
Wheat plant
Brown
G+ive, rod
7
HS1
Leclercia adecarboxylata
MT378327
Humate soil
light green
G-ive, rod
8
HS2
Citrobacter youngae
MT378331
Humate soil
White, dull
G-ive, rod
9
FS2
Enterobacter cloacae
MT872006
Flooded soil
Light green
G-ive, Curve rod
Table 2: Characteristics of antagonistic isolates.
Detection and characterization of siderophores
The development of a yellow to orange colored area on a blue CAS agar plate suggesting siderophore production was reported in 9 isolates. The color shift from blue to orange was caused by siderophores removing Fe from the dye (Figure 2), which was also representative of the hydroxymate form of siderophore [33]. Hydroxymate type of siderophore was also confirmed by the Atkin method. Basically, siderophores are categorized into three categories according to Messenger and Ratledge, 1985 [34]: hydroxamate produced by both bacteria and fungi, and catecholate produced only by bacteria. Table 3 showed the siderophore production index and the siderophore form produced by nine antagonist isolates. For the control of several fungal pathogens that cause plant diseases, siderophores producing bacteria as a biocontrol agent can play a crucial role. [6]. Our results confirm previous research suggesting that siderophore-mediated competition in disease incidence reduction has resulted in pathogen elimination in the rhizosphere [35,36].

Figure 2: Characteristics of antagonistic isolates.
Isolates
Siderophores production index
Types of Siderophore
BR1
2.89±0.73a
Hydroxymate, Catecolate
BR2
1.64±0.11c
Hydroxymate, Catecolate
FS2
1.24±0.08d
Hydroxymate, Catecolate
FS3
1.27±0.06d
Hydroxymate, Catecolate
HS1
1.23±0.08d
Hydroxymate, Catecolate
HS2
1.19±0.04d
Hydroxymate, Catecolate
SF
1.68±0.05bc
Hydroxymate, Catecolate
WM
1.87±0.03c
Hydroxymate, Catecolate
WM2
1.18±0.04d
Hydroxymate, Catecolate
Table 3: Siderophore production index and types of siderophore.
HCN production
HCN productivity of the nine isolates are presented in Figure 3. HCN is a volatile compound which plays a critical role in biological control of some soil-borne diseases [37]. HCN is one of the known biocontrol agent based on its ascribed toxicity against plant pathogens [38,39]. All nine strains showed a positive result of hydrogen cyanide production (Figure 4). Apart from biocontrol properties, biogenic HCN could enhance other plant growth-promoting properties, as well as increase the nutrient availability [38].
Figure 3: Figgal agar method for HCN production assay.
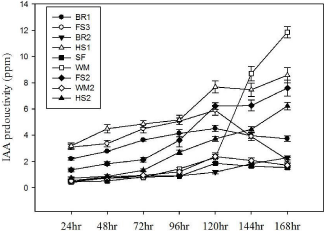
Figure 4: IAA production rate of selected antagonistic isolates.
IAA production
L-tryptophan is commonly found in plant exudates [40]. In the presence of L-tryptophan, it is observed that all the isolated strains tested for IAA biosynthesis were positive. The amount of IAA produced by the nine antagonists varied between different strains at different time intervals (24, 48, 72, 96, 120, 144, 168 hr) when supplemented with L-tryptophan (Figure 4). Most of previous studies have shown that IAA was produced by gram-negative bacteria [41] and few Gram-positive strains are known to produce IAA [42]. In the present study it is shown that five IAA producing strains were Grampositive. Among them, the highest auxin-level producing strain was WM (Pseudomonas alicaligenes), while three strains (HS1, HS2 and FS2) had produced moderate levels of the phytohormone. Microbial IAA production is able to stimulate root growth and influence other developmental processes such as apical dominance, tropical responses, flowering and fruiting [43]. Moreover, it is observed that root rot pathogens could be suppressed by IAA [44]. Since the isolates were capable of producing IAA, it was postulated that the antagonist bacteria developed auxins to enable phytopathogens suppression. However, the production of IAA in vivo by these strains has yet to be determined.
Principal Component Analysis (PCA)
Five variables were used to investigate the impact of related mechanisms on the inhibition of phytopathogen growth by nine antagonist isolates (IAA development, siderophores production index, antagonistic activity against Rhizoctonia solani, antagonistic activity against Phythium sp., and antagonistic activity against Fusarium oxysporum) (Figure 5). The components of the group analysis are made up of nine classes (each representing an antagonist strain). The resulting PCA graph reveals that the first two components (PC1 and PC2) scores account for 66 % of the total variance of the data collection, implying that this statistical study covered more than half of the variables data. The graph shows a close relationship between antagonistic isolates and the metabolites they secrete. The red line arrows refer to the Eigenvectors and display the positive correlations between each factor and variable. Loading scores of the Eigenvector calculated for all variables with values greater than one, indicating a strong correlation between variables. This result supported the hypothesis that the detected inhibition of phytopathogens’ growth was caused by HCN, siderophores, and IAA. The similar inhibitory effect of antifungal activity against three common phytopathogenic fungi indicated that the isolates from this study could represent a new class of antagonistic microorganisms, especially Pseudomonas alicaligenes-WM, which is positively correlated with all variables.
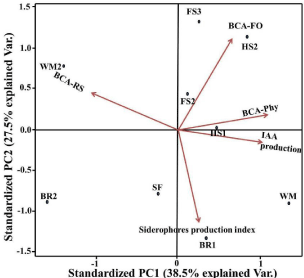
Figure 5: Principal Component Analysis (PCA) plot for Biological control
activity of nine antagonists against three phyopathogens (Rhizoctonia solanii,
Phythium sp., Fusarium oxysporium) and their associated metabolites (HCN,
Siderophores and IAA). BCA-RS=antagonestis activity against Rhizoctonia
solani, BCA-Phy=antagonestis activity against Phythium sp., BCAFO=
antagonestis activity against Fusarium oxysporium.
Formulation for preserved selected antagonist and its viability
Among the nine antagonist microbes, the bacterial strain Pseudomonas alicaligenes-WM was the most effective antagonist strain against fungal pathogens while producing siderophores, HCN and IAA. Therefore, WM strain was used for formulation experiments in search of potential microorganism that could be used as biofertilizer. Among the solid carrier systems, the tablet form is considered to bring and keep the inocula to different places and use it conveniently. It can then be used for mixing with compost or in water as a foliar. The viability of the 12 formulations was tested at 30-day intervals, and the data was summarized in Table 4. The T6 formulation (2.5 g of rice flour, glutinous rice flour, 0.125 g of MSG and 0.02 g of Chitosan) displayed the highest viability of the isolate. This is attributed to the growth control properties of rice flour and glutinous rice flour [45,46], the cryoprotective effect of Monosodium Glutamate (MSG) [47,48] and the antibacterial effect that protect cross-contamination of chitosan [49]. It was found that the survival and cryoprotective effects of the retained formulation decreased as the storage time increased [48]. The formulated T6 can therefore be used as a biofertilizer carrier for further use.
Formulated tablets
Time (Day)
30
60
90
T-1
6x10 7
5x10 5
1x105
T-2
3x10 8
5x10 5
1x103
T-3
1.2x107
4.5x10 5
1x105
T-4
1.1x107
7.5x10 5
7x104
T-5
1.4x107
6.5x10 5
5x104
T-6
5x10 9
1x10 7
1.5x107
T-7
5x10 9
5.5x10 6
-
T-8
1x107
5x10 5
5x104
T-9
1.3x107
1x106
1x101
T-10
1.1x109
2x107
1x104
T-11
1x10 5
1.5x107
5x102
T-12
1x107
1x106
-
Table 4: Viability of isolate’s (Pseudomonas alicaligenes-WM) formulations during storage at room temperature.
Conclusions
Introduction of biocontrol agent to farmlands requires suitable and compatible rhizobateria for the goal of making agriculture more sustainable. Furthermore, it is important that antagonistic compounds developed from soil microbes are investigated to understand how biocontrol bacteria regulate inhibition of pathogens. In this study, the results from in vitro assays suggest that indigenous pseudomonas alcaligens-WM has the potential to protect the host plants from pathogen-induced plant diseases, possibly through the production of siderophores, HCN, IAA production. The formulated carrier can maintain the antagonist microorganisms up to 3 months. Therefore, we recommend that tablet form with our formulation should be the appropriate carrier for further in vivo experiments for biocontrol of soil-borne plant pathogens.
Conflicts of Interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work; there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in this paper.
Funding Statement
AAK, TTW, KTO and PCF are thankful to the financial support by the Research Start-Up Funds from Hainan University in China (KYQD_ZR2017212).
References
- Pahari A, Mishra B. “Characterization of siderophore producing Rhizobacteria and Its effect on growth performance of different vegetables,” International Journal of Current Microbiology and Applied Sciences. 2017; 6: 1398-1405.
- Labuschagne N, Pretorius T, Idris A. “Plant growth promoting rhizobacteria as biocontrol agents against soil-borne plant diseases. Plant growth and health promoting bacteria”, Springer. 2010; 211-230.
- Vinayarani G, Prakash H. “Growth promoting rhizospheric and endophytic bacteria from Curcuma longa L. as biocontrol agents against rhizome rot and leaf blight diseases.” The plant pathology journal. 2018; 34: 218.
- El-Bendary MA. “Bacillus thuringiensis and Bacillus sphaericus biopesticides production.” Journal of basic microbiology. 2006; 46: 158-170.
- Hallmann J, Davies KG, Sikora R. “Biological Control Using Microbial Pathogens, Endophytes and Antagonists.” Root-knot nematodes. 2009; 380.
- Patil S, Bheemaraddi MC, Shivannavar CT, Gaddad SM. “Biocontrol activity of siderophore producing Bacillus subtilis CTS-G24 against wilt and dry root rot causing fungi in chickpea.” Journal of Agriculture and Veterinary Sciences. 2014; 07: 63-68.
- Whipps JM. “Microbial interactions and biocontrol in the rhizosphere.” Journal of experimental Botany. 2001; 52: 487-511.
- Lugtenberg B, Kamilova F. “Plant-growth-promoting rhizobacteria,” Annual review of microbiology. 2009; 6: 541-556.
- J. M. Raaijmakers, T. C. Paulitz, C. Steinberg, C. Alabouvette and Y. Moënne-Loccoz “The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms,” Plant and soil, vol. 321, no. (1-2), pp. 341-361, 2009.
- Sadi T, Rahim N, Rashdi A, Nejis N, Hassan R. “Bio prospecting and management of microorganisms.” National conference on agro biodiversity conservation and sustainable utilization. 2006; 129-130.
- Umi K, Sariah M. “Utilization of microbes for sustainable agriculture in Malaysia: current status. Bio prospecting and management of microorganisms.” National Conference on Agro biodiversity conservation, and sustainable utilization. 2006; 27-29.
- Sharma A, Johri B. “Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions.” Microbiological research. 2003; 158: 243-248.
- Patel K, Ahire J, Pawar SP, Chaudhari BL, Shouche YS, Chincholkar SB. “Evaluation of probiotic characteristics of siderophoregenic Bacillus spp. isolated from dairy waste.” Applied biochemistry and biotechnology. 2010; 160: 140.
- Spaepen S, Vanderleyden J, Remans R. “Indole-3-acetic acid in microbial and microorganism-plant signalling.” FEMS microbiology reviews. 2007; 31: 425-448.
- Santner LI, Calderon-Villalobos A, Estelle M. “Plant hormones are versatile chemical regulators of plant growth.” Nature chemical biology. 2009; 5: 301- 307.
- Ramette M, Frapolli G, Défago A, Moënne-Loccoz Y. “Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability.” Molecular Plant-Microbe Interactions. 2003; 16: 525-535.
- Michelsen F, Stougaard P. “Hydrogen cyanide synthesis and antifungal activity of the biocontrol strain Pseudomonas fluorescens In5 from Greenland is highly dependent on growth medium,” Canadian journal of microbiology. 2012; 58: 381-390.
- Pal K, Tilak K, Saxena A, Dey R, Singh C. “Antifungal characteristics of a fluorescent Pseudomonas strain involved in the biological control of Rhizoctonia solani.” Microbiological research. 2000; 155: 233-242.
- Nagarajkumar M, Bhaskaran R, Velazhahan R. “Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen,” Microbiological Research. 2004; 159: 73-81.
- Rezzonico F, Zala M, Keel C, Duffy B, Moënne-Loccoz Y, Défago G. “Is the ability of biocontrol fluorescent pseudomonads to produce the antifungal metabolite 2, 4-diacetylphloroglucinol really synonymous with higher plant protection?,” New Phytologist. 2007; 173: 861-872.
- Insunza V, Alström S, Eriksson K. “Root bacteria from nematicidal plants and their biocontrol potential against trichodorid nematodes in potato.” Plant and Soil. 2002; 241: 271-278.
- Gallagher LA, Manoil C. “Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning.” Journal of bacteriology. 2001; 183: 6207- 6214.
- Uzair R, Kausar SA, Bano S, Fatima S, Badshah M, Habiba U, et al. “Isolation and molecular characterization of a model antagonistic Pseudomonas aeruginosa divulging in vitro plant growth promoting characteristics.” BioMed research international. 2018.
- R Coico R. “Gram staining.” Current protocols in microbiology. 2006; 1.
- Schwyn A, Neilands J. “Universal chemical assay for the detection and determination of siderophores,” Analytical biochemistry. 1987; 160: 47-56.
- Silva-Stenico ME, Pacheco FTH, Rodrigues JLM, Carrilho E, Tsai SM. “Growth and siderophore production of Xylella fastidiosa under iron-limited conditions,” Microbiological Research. 2005; 160: 429-436.
- Pal R, Gokarn K. “Siderophores and pathogenecity of microorganisms.” Journal of Biosciences and Technology. 2010; 1: 127-134.
- Arnow LE. “Colorimetric determination of the components of 3, 4-dihydroxyphenylalanine-tyrosine mixtures.” Journal of Biological Chemistry. 1937; 118: 531-537.
- Atkin J, Neilands A, Phaff H. “Rhodotorulic acid from species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a new alanine-containing ferrichrome from Cryptococcus melibiosum.” Journal of Bacteriology. 1970; 103: 722-733.
- Ehmann A. “The Van Urk-Salkowski reagent-a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives.” Journal of Chromatography A. 1977; 132: 267-276.
- García-Bayona L, Comstock LE. “Bacterial antagonism in host-associated microbial communities,” Science. 2018; 361: 6408.
- Földes T, Banhegyi I, Herpai Z, Varga L, Szigeti J. “Isolation of Bacillus strains from the rhizosphere of cereals and in vitro screening for antagonism against phytopathogenic, food-borne pathogenic and spoilage micro-organisms.” Journal of applied microbiology. 2000; 89: 840-846.
- Milagres M, Machuca A, Napoleao D. “Detection of siderophore production from several fungi and bacteria by a modification of Chrome Azurol S (CAS) agar plate assay,” Journal of Microbiological Methods. 1999; 37: 1-6.
- Messenger JM, Ratledge C, Moo-Young M. “Siderophores,” Comprehensive Biotechnology, Pergamon, New York. 1985; 3: 275-295.
- Gupta P, Maheshwari DR. “ Plant growth enhancement and suppression Macrophomina phaseolina causing charcoal rot of peanut by fluorescent Pseudomonas,” Biological Fertility of Soils. 2002; 35: 399-405.
- Chandra S, Dubey RC, Maheskwari DK. “Rhizosphere competent Mesorhizobium loti MP 6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris).” Brazilian Journal of Microbiology. 2004; 24: 35-39.
- Haas A, Défago G. “Biological control of soil-borne pathogens by fluorescent pseudomonads,” Nature reviews microbiology. 2005; 3: 307-319.
- Tomaz Rijavec L. “Hydrogen cyanide in the rhizosphere: not supressing plant pathogens, but rather regulating the availability of phosphate.” Frontiers in microbiology. 2016; 7: 1785.
- Heydari A, Pessarakli M. “A review on biological control of fungal plant pathogens using microbial antagonists,” Journal of biological sciences. 2010; 10: 273-290.
- Hardoim PR, van LS, Overbeek A, van Elsas JD. “Properties of bacterial endophytes and their proposed role in plant growth.” Trends in microbiology. 2008; 16: 463-471.
- Datta A, Basu P. “Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanus cajan.” Microbiological research. 2000; 155: 123-127.
- Wahyudi T, Astuti RP, Widyawati A, Mery A, Nawangsih AA. “Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting rhizobacteria,” Journal of Microbiology and Antimicrobials. 2011; 3: 34-40.
- Spaepen S. “Plant hormones produced by microbes. Principles of plantmicrobe interactions.” Springer. 2015; 247-256.
- Khare A, Arora NK. “Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea,” Current microbiology. 2010; 61: 64-68.
- Choi BK, Park SY, Ha SD, Kum JS, Lee HY, Park JD. “Effects of Storage Conditions of Rice Flour on Growth Properties of Natural Microflora.” Journal of the Korean Society of Food Science and Nutrition. 2007; 36: 921-925.
- Ji Y, Zhu K, Qian H, Zhou H. “Microbiological characteristics of cake prepared from rice flour and sticky rice flour.” Food Control. 2007; 18: 1507-1511.
- Seguro K, Tamiya T, Tsuchiya T, Matsumoto J. “Cryoprotective effect of sodium glutamate and lysine-HCl on freeze denaturation of lactate dehydrogenase.” Cryobiology. 1990; 27: 70-79.
- Coulibaly R, Dubois-Dauphin J, Destain M-L, Fauconnier G, Lognay A, Thonart P. “The resistance to freeze-drying and to storage was determined as the cellular ability to recover its survival rate and acidification activity.” International Journal of Microbiology. 2010.
- Atay HY. “Antibacterial Activity of Chitosan-Based Systems.” Functional Chitosan. 2019; 457-489.