
Research Article
Austin J Biotechnol Bioeng. 2023; 10(1): 1121.
Purification and Characterization of Glucosamine Oxidase from Aspergillus Terreus NUK-1204 for Production of Glucosaminic Acid
Shuen-Fuh Lin*; Pang-Hung Huang
Department of Life Sciences, National University of Kaohsiung, 700, Kaohsiung University Road, Taiwan
*Corresponding author: Shuen-Fuh Lin Department of Life Sciences, National University of Kaohsiung, 700, Kaohsiung University Road, Nanzih District, Kaohsiung, 811, Taiwan. Tel: +886 7 5919227; Fax: +886 7 5919404 Email: sflin@nuk.edu.tw
Received: August 21, 2023 Accepted: September 16, 2023 Published: September 23, 2023
Abstract
D-Glucosaminic acid (2-amino-2-deoxy-D-gluconic acid) is a valuable glucosamine derivative from the oxidation of the glucosamine (2-amino-2-deoxy-D-glucose). It can be produced via the fermentation of certain microorganisms, which involves complicated purification task, though. In an attempt to find an enzyme suited to glucosaminic acid production, the study presented a purified homogeneous glucosamine oxidase deriving from the wheat bran culture of a soil-isolated fungal strain, Aspergillus terreus NUK-1204. Maximum activity was observed in the wheat bran solid culture after five days of NUK-1204 growth, following its release from cells at 0.011 unitmL -1culture filtrate. The new kind of oxidase comprises a single polypeptide chain with a molecular mass of 54 kDa, containing FAD and exhibiting absorption maxima at 274, 372 and 439 nm. The enzyme was stable in 4.5-11 pH range, with an optimal reaction pH at 10.5 and optimal reaction temperature at 30°C. It remained stable at 20-50°C for 1 hr. The relative activity of glucosamine oxidase towards glucosamine and N-acetyl-glucosamine was 100:7, demonstrating high substrate specificity. It doesn’t inhibit glucosamine. To the best of our knowledge, this is the first report ever on the discovery of a glucosamine oxidase based on enzyme substrate specificity.
Keywords: Glucosamine oxidase; Aspergillus terreus; Glucosaminic acid
Introduction
There have been studies on the biosynthesis of D-Glucosaminic acid (2-amino-2-deoxy-D-gluconic acid), a component of bacterial lipopolysaccharides with species of a-amino acid, most involving the oxidation of D-glucosamine (2-amino-2-deoxy-D-glucose) via a specific membrane oxidase in the bacteria [1]. While knowledge on this enzyme is still limited, D-glucosaminic acid has been used as sweetener, condiments, and chiral synthon [2]. It also belongs to a chiral pool and is a trigger for the asymmetric synthesis of various amino acids, such as 4,5,6-trihydroxynorleucine [3], and several glycosidase inhibitors, like (+)-castanosporspermine [4], plus an expanding scope of potential applications. Carbohydrate oxidation has been widely applied in pharmaceutical production as biocatalyst and diagnostic reagent. Given the key role of regioselectivity and steroselectivity in these reactions, it is critical to find optimal biocatalysts [5,6]. Various sugar oxidoreductases have been identified from a variety of sources, including bacteria [7], fungi [8,9], and algae [10,11], most of which being active with mono and disaccharides primarily, with only a few exhibiting high specificity toward amino-sugar. One example is N-acyl-D-hexosamine oxidase obtained from Pseudomonas sp. [12], which shows high reactivity to N-acyl-D-hexosamines containing N-acyl-D-glucosamine and N-acyl-D-galactosamine. In contrast, D-glucosamine and D-glucose demonstrated lower activity to the enzyme.
In line with the proposal of Danneel et al. [13], the study applied a rapid and convenient screening method for the in situ assay of sugar oxidase-producing soil fungi, on top of under solid-state fermentation and the peroxidase-chromogen method for assaying oxidase activity. Strain NUK-21, identified as Myrmecridium flexuosum, was isolated and named lactose oxidase, based on its substrate specificity [14]. Fortunately, strain NUK-1204, identified as Aspergillus terreus, was isolated. Oxidase deriving from the purification of the aqueous extract of wheat bran culture of this strain showed high reactivity toward D-glucosamine, while D-N-acetylglucosamine and D-glucose exhibited lower activity. The new kind of oxidase was named glucosamine oxidase, based on its substrate specificity. Its purification and some properties follow.
Materials and Methods
Chemicals
Fractogel DEAE-650M, HW-55 gel filtration resins were purchased from Merck (Darmstadt, Germany). Toyopearl Phenyl-650M was purchased from Toyo Soda Manufacturing (Tokyo, Japan). Ultragel-HA was acquired from IBF Biotechnics (Pairs, France). Protein assay dye and SDS-PAGE molecular weight standards were obtained from Bio-Rad Laboratories (Hercules, CA, USA). Sephacryl S-200 HR16/60 FPLC column was from GE Healthcare Bio-Science (Uppsala, Sweden). FPLC molecular weight standards, 4-aminoantipyrine (4AA), peroxidase and all the sugars were from Sigma Chemical Co. (St. Louis, MO, USA). All other chemicals were of analytic reagent grade.
Microorganisms and Culture Conditions
The fungal strain NUK-1204 was isolated from the soil, identified as Aspergillus terreus and deposited at the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan) of Taiwan under accession number BCRC 930228. The organism was maintained on YM agar slants (1% glucose, 0.5% peptone, 0.3% malt extract, and 0.3% yeast extract) at 30°C.
Production of Glucosamine Oxidase
The NUK-1204 strain produced glucosamine oxidase activity on wheat bran culture. For enzyme production, this strain was taken from a slant culture and inoculated in 250 ml flasks, each of which contained 5g of wheat bran supplemented with 17.5 ml of water. After 5 days of cultivation at 30°C, the entire culture was used for enzyme preparation.
Enzyme Assay
The enzyme activity was assayed through the peroxidase-4AA method. An aliquot of enzyme was incubated at 30°C in 1 mL of 50 mM Tris–HCl buffer (pH 7.8, buffer A) containing 50 mM glucosamine, 1 U peroxidase, 0.1 mM 4AA, and 1 mM phenol. The increase in optical density was measured at 500 nm for 1 min. The enzyme activity was estimated by monitoring the consumption of oxygen with an oxygraph (Oxy-5, Gilson Medical Electronic, France). The reaction was initiated by the addition of appropriate amounts of the enzyme to the reaction mixture containing 25 mM glucosamine in buffer A, and the initial velocity of oxygen consumption was measured. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmolmin-1 of H2O2 or consumed 1 μmol min-1 of O2 at 30°C.
Enzyme Purification
A typical purification scheme of the glucosamine oxidase from wheat bran culture is described as follows. All operations were conducted at 4°C.
Preparation of Crude Extract
The wheat bran culture of strain NUK-1204 (120g) was soaked in 1 l of 20 mM borate buffer (pH 9.5) with 0.1% triton X-100 for 30 min and squeezed through a fine-mesh cloth. The aqueous extract was then centrifuged at 8000g for 30 min to remove particles.
Ammonium Sulfate Fractionation
The clarified aqueous extract (830 mL) was brought to 65% saturation with ammonium sulfate; the precipitate was removed via centrifugation at 9,000×g at 4°C for 30 min. The resulting filtrate was collected via centrifugation.
Toyopearl Phenyl-650M Column Chromatography
The 65% saturated ammonium sulfate filtrate was applied to a Toyopearl phenyl-650M column (2.5×30 cm) (Toyo Soda Manufacturing), which was pre-equilibrated with buffer A containing 65% saturated ammonium sulfate. Glucosamine oxidase was eluted with a 1,400 mL linear gradient of 65–0% ammonium sulfate in buffer A at a flow rate of 2.5 ml/min.
Fractogel HW-55 Column Chromatography
The enzyme solution was applied to a Fractogel HW-55 column (2.5cm×115cm), which was pre-equilibrated with 50 mM buffer A at a flow rate of 0.25 ml/min. To concentrate the pooled active fractions (352 mL), ammonium sulfate was added to the enzyme solution to a final concentration of 2M and then recharged to a small Toyopearl phenyl-650M column (1cm×6 cm), which was pre-equilibrated with buffer A containing 2M ammonium sulfate. In this process, the enzyme was eluted directly with buffer A. The volume of pooled active fractions was 15 mL.
Fractogel DMAE-650 Column Chromatography
The enzyme solution (70 ml) was applied to a Fractogel DMAE-650 column (2.5cm×30 cm), which was pre-equilibrated with buffer A. The enzyme was eluted by linear gradient from 0 to 0.15 M NaCl in buffer A at a flow rate of 2.0 ml/min.
Ultragel-HA Hydroxylapatite Column Chromatography
The pooled active fractions were further purified by applying them to an Ultragel-HA hydroxylapatite column (2.5cm×15 cm), which was pre-equilibrated with 10 mM phosphate buffer (pH 7.0). The enzyme was eluted with a 600-mL linear gradient of 10–150 mM phosphate buffer at a flow rate of 1.5 ml/min. The active fractions were pooled and stored at -20°C.
Amino-Terminal Amino Acid Sequence of Glucosamine Oxidase
Amino acid sequence of the purified peptide (0.01mg) was determined by an automated Edman degradation with an Applied Biosystems, Procise 494 protein sequencer (Foster City, CA, USA).
Other Analytical Methods
The molecular mass of the native enzyme was estimated through gel filtration under the following conditions: system, Fast Performance Liquid Chromatography (FPLC) system; pump, P-500; controller, LCC-500; detection, absorbance at 280 nm; column, Sephacryl S-200 HR16/60 FPLC column; solvent, 100 mM NaCl in 10 mM acetate buffer (pH 5.5). The molecular mass of the denatured enzyme was determined through SDS polyacrylamide gel electrophoresis (SDS-PAGE) on 10% acrylamide slabs using a modified Laemmli buffer system [15]. Protein concentrations were determined through the Bradford method with bovine serum albumin as the standard [16]. Protein assay reagents were purchased from Bio-Rad. The Mass spectrometry data were obtained using APCI ion source from Advion expression® CMS instrument (Advion Inc., Ithaca, NY, USA). Nuclear magnetic resonance spectra were recorded on a Varian-Mercury plus 300 MHz spectrometry (Agilent, Santa Clara, CA, USA). 300 MHz for 1H and 75 MHz for 13C with D2O (deuterium oxide) as the solvent.
Thin-Layer Chromatography (TLC) Analysis of the Reaction Product
A reaction mixture (1mL) containing 2.5% glucosamine (w/v), 2 units of glucosamine oxidase, and 10 units of catalase was incubated at 30°C with a reciprocal shaker (120 rpm). Subsequently, the reaction product was detected using Thin-Layer Chromatography (TLC) with silica gel 60 plates (Merck, Germany). Aliquots (approximately 1μL) of sample were applied to the TLC plate and developed in n-butanol/methanol/ammonium water/water (5:4:2:1, v/v). The plate was sprayed with 50% sulfuric acid dissolved in ethanol /water (1:1 v/v) and then heated at 150°C until charring occurred.
Results
Enzyme Production
Extracellular glucosamine oxidase deriving from Aspergillus terreus NUK-1204 was produced in Solid-State Fermentation (SSF) only, rather than submerged culture. Compared with rice bran, corn flour, sugarcane bagasse, and soy meal, what bran was found to be the best medium for enzyme production, as with application of wheat bran as a substrate in SSF, the addition of other carbon sources (e.g., glucose, fructose, glycerol, starch, and cellulose) or nitrogen sources (e.g., peptone, yeast extract, malt extract, and urea) didn't enhance enzyme production. A mixture of wheat bran and water in the ration of 1:3.5 (w/v) was found to be the optimal fermentation medium. In enzyme product, it typically took five days at 30o to reach maximum glucosamine oxidase activity (Figure 1). About 0.011 U of enzyme activity per mL could be extracted from the culture broth with 20 mM borate buffer (pH 9.5) containing 0.1% triton X-100.
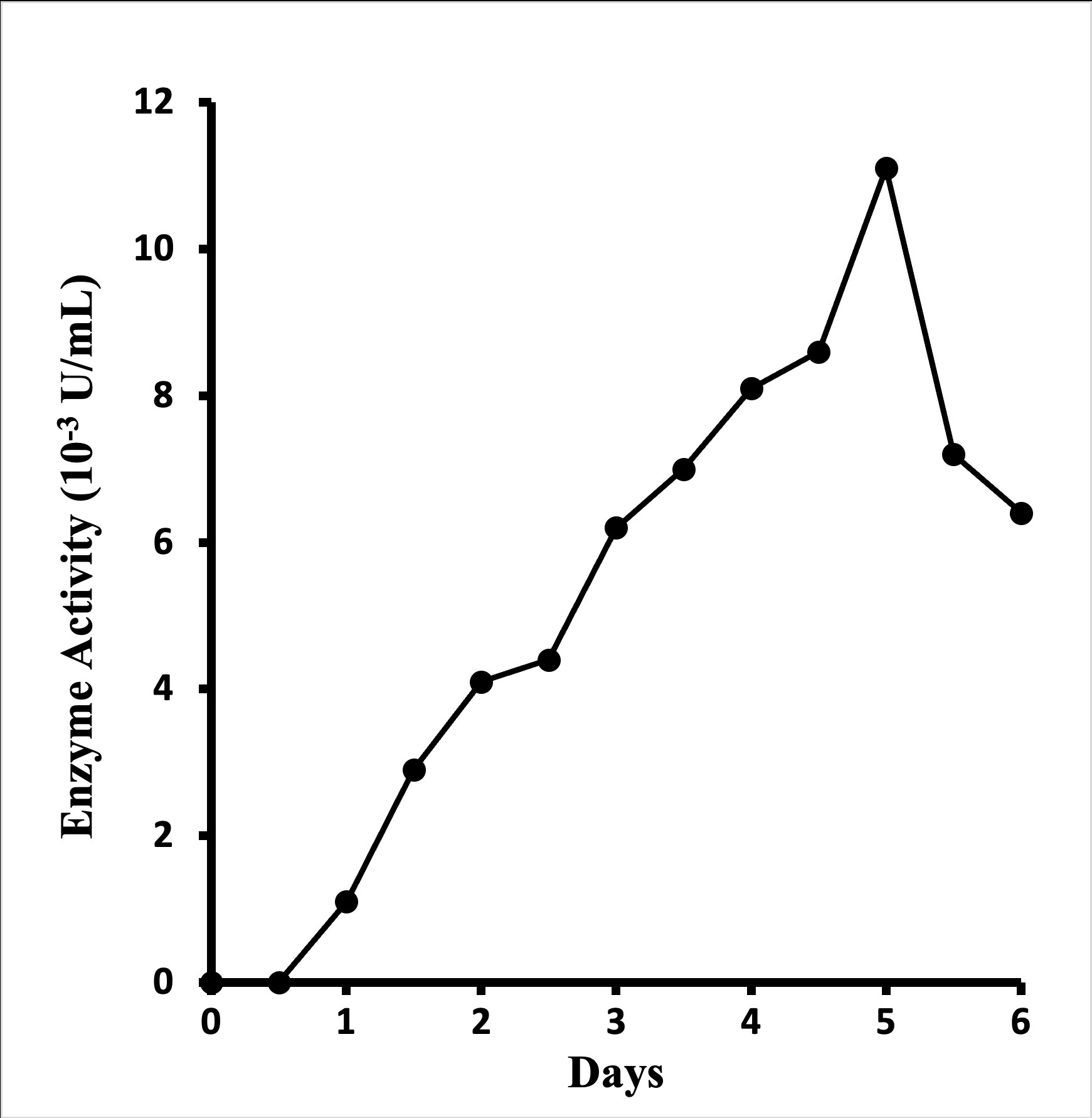
Figure 1: Time course of enzyme production by Aspergillus terreus NUK-1204 in wheat-bran solid-state fermentation at 30o.
Purification of Glucosamine Oxidase
Table 1 on the purification for glucosamine oxidase shows a 101-fold increase in the enzyme's specific activity and 36% recovery in total activity. The purity of the enzyme was confirmed by electrophoresis and exhibited a single band in SDS-PAGE (Figure 2). The specific activity of the purified enzyme was 1.1 U mg-1 of protein.
Step
Volume
(ml)Total protein
(mg)Total activitya (unit)
Specific activitya
(U/mg)Yield (%)
Purification (fold)
Crude extract
830
816
9.13
0.011
100
1
(NH4)2SO4 precipitation
1040
307
5.69
0.019
62
2
Toyopearl phenyl-650M
352
134
7.78
0.058
85
5
HW-55
70
55
6.58
0.120
72
11
DEAE-650M
575
14
3.82
0.273
42
25
Ultrogel-HA
176
3
3.32
1.107
36
101
Table 1: Purification of glucosamine oxidase from the A. terreus NUK-1204 strain.
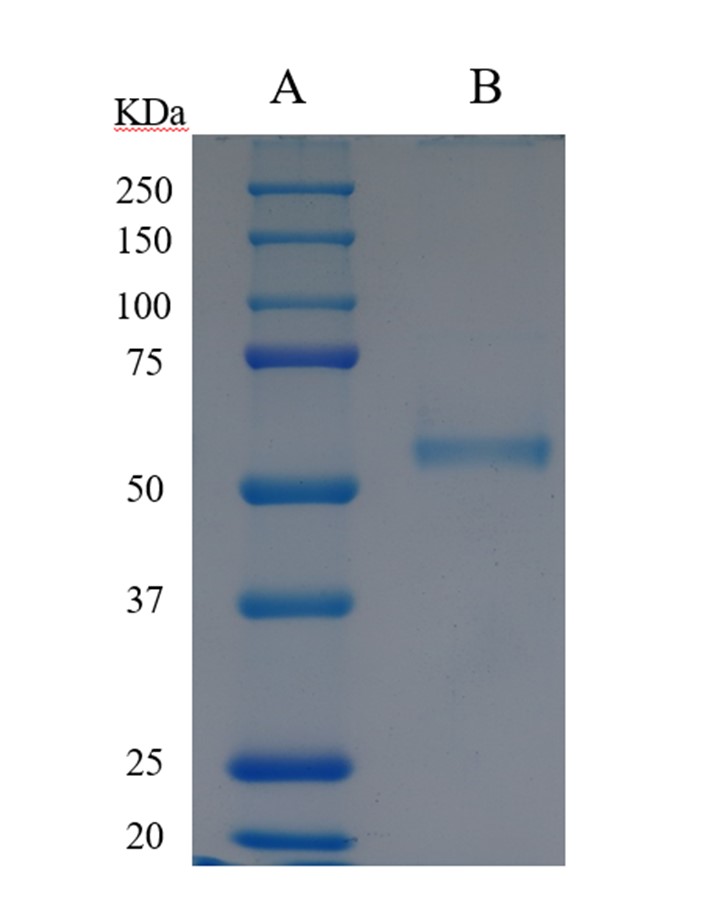
Figure 2: SDS-PAGE analysis of glucosamine oxidase. The gel concentration was 10%. Lane A, molecular mass markers. Lane B, 10 μg of the purified enzyme.
Molecular Weight
The purified enzyme was shown to be homogenous, as shown by analysis via SDS-PAGE (Figure 2). The enzyme's molecular mass was estimated at 54 kDa via SDS-PAGE (Figure 2) and size-exclusion FPLC on a Sephacryl S-200 HR16/60 column (data not shown), indicating that the enzyme comprises a single polypeptide chain.
Amino-Terminal Amino Acid Sequence Analysis
The N-terminal amino acid sequence of the glucosamine oxidase's first 21 residues was RDA LLPSSLQTCLRNTGAKIVY. Via the use of BLASTP program, no significant homology sequence was found in the GenBank database.
Absorption Spectrum
Figure 3 shows the absorption spectra of purified glucosamine oxidase in oxidized and reduced states. The enzyme's absorption peaks at 276 nm, 372 nm, and 439 nm, which could also be observed in FAD. With addition of glucosamine, the FAD prosthetic group in the enzyme changed from an oxidized state to a reduced state, causing the absorption peak of the enzyme at 439 nm to disappear (Figure 3-B). These spectral profiles suggest that the enzyme is a flavoprotein containing FAD. value of this enzyme was calculated to be 15.9.
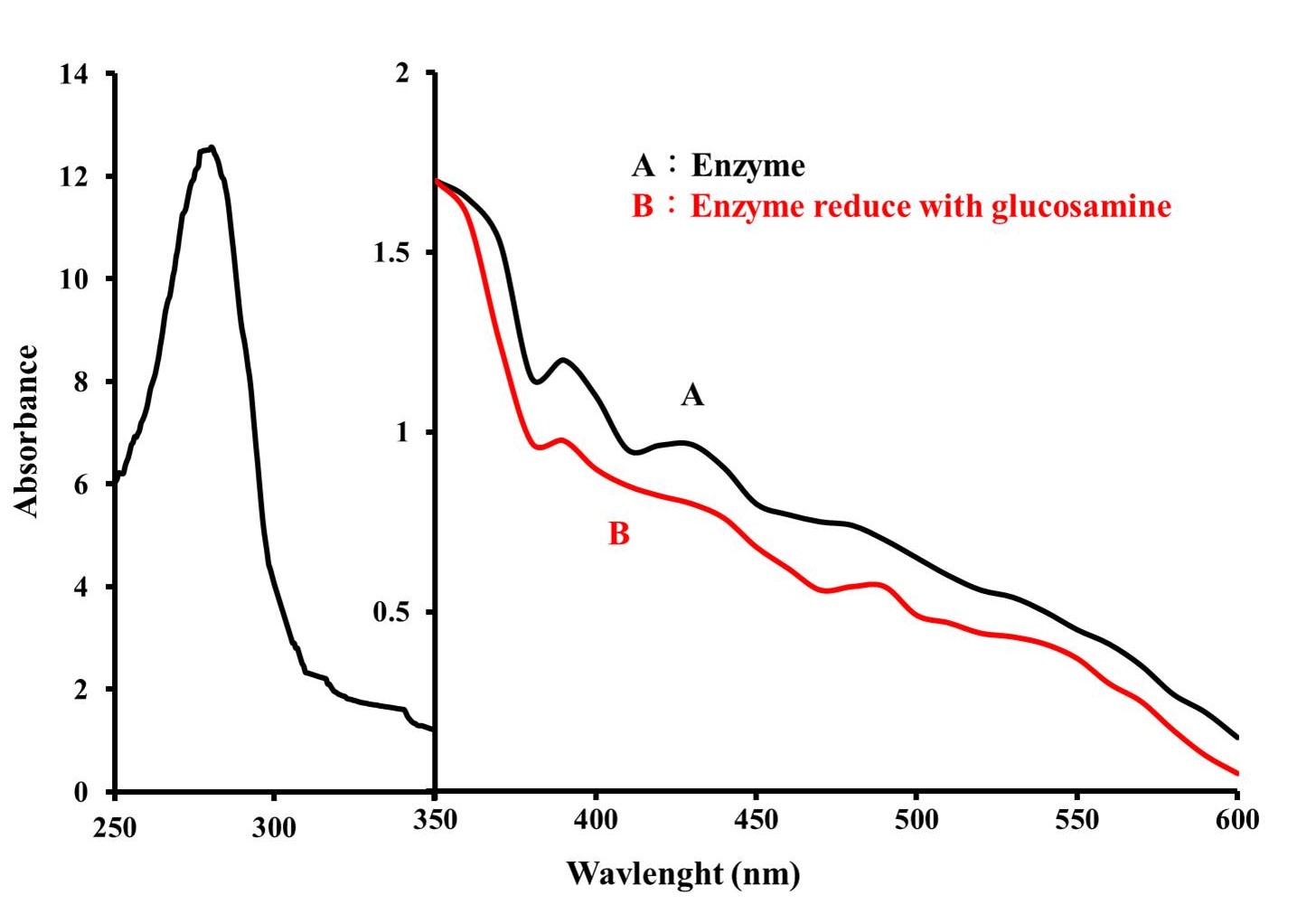
Figure 3: Absorption spectrum of glucosamine oxidase (3mg/mL) in buffer A. Absorption spectrum was taken with a NanoDrop One/One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific). (A) Native enzyme, (B) The prosthetic group of enzyme was reduced by adding glucosamine to a concentration of 200 mM.
Optimal Reaction pH and pH Stability
The oxygraph method was used to examine the enzyme activity's pH dependency. The bell-shaped pH versus activity profile in Figure 4-A shows that the enzyme's optimal reaction pH stands at around 10.5. The enzyme was incubated at 30°C for 1 h under various pH conditions to observe its pH stability. Figure 4-B shows that the enzyme activity measured through peroxidase-4AA method is quite stable in the pH range from 4.5 to 11.
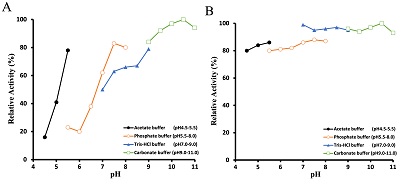
Figure 2: Effects of pH on the activity and stability of glucosamine oxidase. The optimal pH of glucosamine oxidase (A) was determined at the pH values indicated by oxygraph method. The stability of glucosamine oxidase (B) was assayed by incubating the enzyme at the defined pH at 30°C. After 1 h, the residual activity was estimated at pH 7.8 using the peroxidase-4AA method, as described in the text.
Optimal Reaction Temperature and Thermal Stability
As shown in Figure 5-A, the optimal temperature for glucosamine oxidation under the standard oxygraph conditions was observed to be 30°C. The effect of temperature on the stability of the enzyme was examined. As shown in Figure 5-B, the enzyme remained stable up to 50°C.
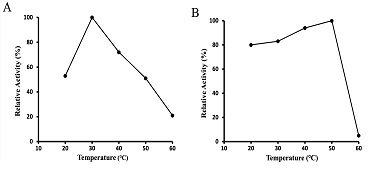
Figure 5: Effects of temperature on activity and stability of glucosamine oxidase. The optimal temperature of glucosamine oxidase (A) was determined at the temperature values indicated by oxygraph method. The stability of glucosamine oxidase (B) was assayed by incubating the enzyme in the defined temperature at 30°C. After 1 h, the residual activity was estimated at pH 7.8 using the peroxidase-4AA method.
Substrate Specificity
The study looked into the ability of NUK-1204 oxidase to oxidize various sugars in buffer A with the peroxidase-4AA method (Table 2). NUK-1204's oxidase demonstrated more potent activity in glucosamine than in N-acetyl-glucosamine. Apparently, only glucosamine and N-acetyl-glucosamine (glucosamine; which is NH2 group at the C2 position) are acceptable substrates. No significant activity could be observed in all the other carbohydrates tested. The study named the enzyme glucosamine oxidase, as it demonstrated the highest catalytic efficiency with glucosamine. To the best of our knowledge, it is the first report ever on the discovery of a glucosamine based on enzyme substrate specificity. Moreover, the study estimated the Km and kcat values of the glucosamine substrates with Hanes-Woolf plots, finding that the apparent Km values of the glucosamine was 34 mM and the Kcat value was 0.42 s-1. Thee Kcat/Km value was 12.35 s-1mM-1.
Substrates
Relative activity (%)
Glucosamine
100
N-acetylglucosamine
6.5
Glucose
Ribose0
0
Fructose
0
Arabinose
0
Xylose
0
Galactose
0
Mannose
0
Sorbitol
0
Mannitol
0
Xylitol
0
Maltose
0
Lactose
0
Cellobiose
0
Enzyme activity was assayed by measuring peroxidas-4AA method as described in the text.
Table 2: Substrate specificity of glucosamine oxidase.
Effect of Metals
Effect of various metals on the glucosamine oxidase activity were investigated. As shown in Table 3, most of the metal ions had little effect on NUK-1204 oxidase activity, but Cu 2+and Fe2+ both inhibited enzyme activities, reducing them to 73% and 77%, respectively. Glucosamine oxidase activity was increased by Ni2+ and Hg2+to 120% and 116%, respectively. While Hg2+acts as an inhibitor for most enzymes, it increased the activity of glucosamine oxidase by 16%.
Metal ions (1 mM)
Relative activity (%)
Control
100
AgNO3
82
Al2(SO4)3
104
CaCl2
59
CdCl2
99
CoCl2
112
CuSO4
73
FeSO4
77
HgCl2
116
MgSO4
108
MnSO4
110
NiCl2
120
PbCl2
98
SnCl2
91
The residual enzyme activity was assayed by Oxygraph method as described in the text. Glucosamine oxidase (5 μg) was incubated in buffer A containing various metal ions for 1h at 30°C.
Table 3: Effect of metal ions on glucosamine oxidase activity.
Effect of Chemicals
As shown in Table 4, while most of the chemicals had little effect on NUK-1204 oxidase activity, N-ethylmaleimide, 10-phenanthroline, and 2-bromo-4'-nitroacetophenol, inhibited the enzyme's activities, reducing them to 79%, 74% and 67%, respectively. 2-bromo-4'-nitroacetophenol and N-ethylmaleimide are modifiers of histidine and cysteine respectively, and 1, 10-phenanthroline is a chelating agent of iron ions, suggesting their possible involvement in the catalytic mechanism for the enzyme.
Chemicals
Relative activity (%)
Control group
100
2,4'-dibromoacetophenone
81
2-bromo-4'-nitroacetophenol
79
NaN3
84
EDTA
84
N-ethylmaleimide
74
1,10-phenanthroline
67
H2O2
95
PMSF
95
The residual enzyme activity was assayed by oxygraph method as described in the text. Glucosamine oxidase (5μg) was incubated in buffer A containing various chemicals for 1h at 30°C.
Table 4: Effect of chemicals on glucosamine oxidase activity.
Reaction Product
The reaction product of NUK-1204 oxidase started with glucosamine as the substrate. After the aerobic incubation of glucosamine with an excess amount of NUK-1204 oxidase in catalase at 30°C for 3h, the reaction products isolated from the reaction mixture was identical with the authentic glucosaminic acid on TLC assayed by silica gel 60 plates. Only one new compound was found with the same Rf value as glucosaminic acid (Figure 6). It was then purified via high-performance liquid chromatography was done at 60o with a Beckmanμ-Spherogel column (8.0mm×300mm) and deionized water as a eluate. The negative ion electrospray ionization-mass spectrometry was measured and it exhibited a [M-H]-1 ion at m/z 194.1, with a molecular weight of 195 Da, equal to the glucosaminic acid standard. The 13C nuclear magnetic resonance spectrum showed that the chemical shift of the carboxylic acid carbon (C1) is 172.19 ppm (Figure 7), confirming that the glucosamine oxidase reaction product was glucosaminic acid. To summary, it is concluded that glucosamine oxidase catalyzed the following reaction.

Figure 6: TLC analysis of the product. Lane A, glucosamine; lane B, complete conversion product from glucosamine; lane C, standard of glucosaminic acid.
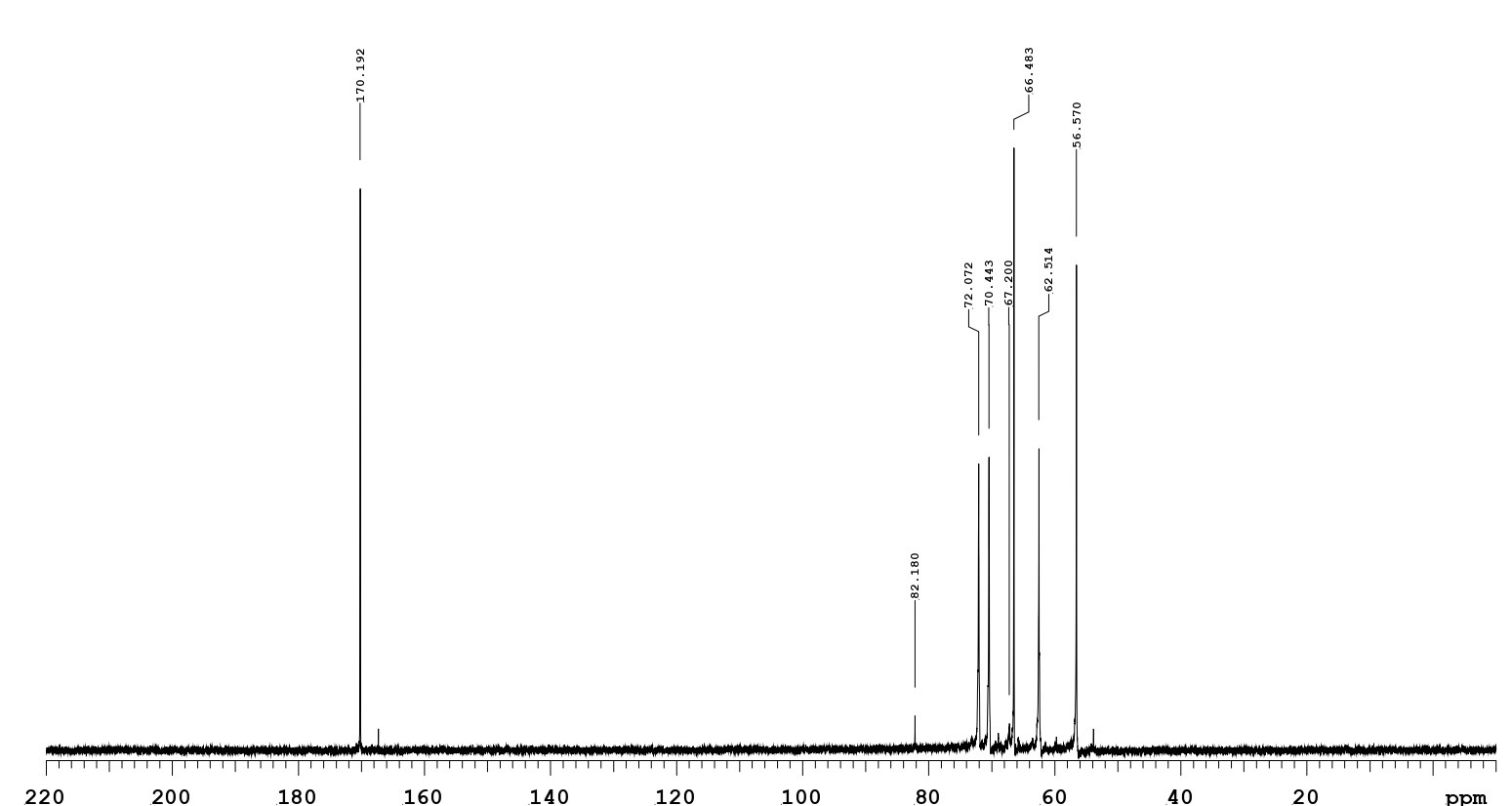
Figure 7: The 13C-NMR spectrum (75 MHz, D2O) of a glucosaminic acid product by glucosamine oxidase.
Glucosamine + O2 → Glucosaminic acid+H2O2
Discussion
Solid-state fermentation refers to the growth of microorganism on the surface of solid substrate with low moisture, similar to its native habitat [17], which is conducive to enzyme growth. The study employed a selective and rapid in situ assay in detecting H2O2-producing oxidase deriving from soil fungi in wheat-bran solid-state culture of various fungi strains, such as glucooligosaccharide oxidase of Acremonium strictum [18], glycerol oxidase of Penicillium TS-622 [19], lactose oxidase of Myrmecridium flexuosum [14] and glucosamine oxidase of Aspergillus terreus NUK-1204. Many of these oxidases, including NUK-1204 oxidase, could be produced in wheat-bran solid state culture only, rather than submerged culture.
Judging from its substrate specificity, as shown in Table 2, the glucosamine oxidase purified from the wheat bran culture of Aspergillus terreus NUK-1204 is indeed a new type of oxidase, outperforming known sugar oxidases in reactivity towards glucosamine, the favored substrate. Moreover, the glucosamine oxidase boasts N-terminal amino acid sequence with no match in homology sequence in literature. The study is believed to be the first report ever on a new glucosamine oxidase.
Table 5 presents a summary of the general properties of glucosamine oxidase deriving from A. terreus NUK-1204, N-acetyl glucosamine oxidase from seudomonas sp. 15-1[12], vanillyl-alcohol oxidase from Fusariium garminearum [19], a glucose oxidase from A. niger [20], with all the four enzymes containing FAD. While glucosamine oxidase and vanillyl-alcohol oxidase are monomeric proteins, n-acetyl glucosamine oxidase and glucose oxidase both comprise two identical and four non-identical subunits, with enzyme molecular weights ranging from 60 to 170 KDa. The molecular weight of the enzyme in the study stands at 54 KDa.
Enzyme
Strain
Mw (kDa)
Prosthetic group
Optimal
pHSubstrate specific
Glucosamine
Glucose
N-acetyl glucosamine
Glucosamine oxidase
A. terreus
NUK-120454
FAD
10.5
100
0
6.5
N-acetyl glucosamine oxidase
Pseudomonas sp.
170
FAD
8.0
14
-
100
Vanillyl-alcohol
oxidaseFusariium garminearum
60
FAD
7.6
-
0.5
100
Glucose oxidase
A. niger
160
FAD
5.6
2
100
-
Table 5: Comparison of the properties of various sugar oxidoreductases.
A most surprising characteristic of this enzyme is its high optimal reaction pH (pH 10.5), a far cry from other oxidases, all of which have their optimal reaction pHs in a weak acidic (pH 5.6) and basic range (pH 7.6 and 8.0). Another outstanding characteristic of the enzyme in the study is its substrate specificity, as it exhibited high activity toward glucosamine, compared with the low selectivity of the other three oxidases towards glucosamine. So far, no other known oxidative enzymes can use glucosamine as the optimal reaction substrate. It is evident that the glucosamine oxidase deriving from NUK-1204 is a new type of oxidative enzyme.
Conclusion
The study presents a novel glucosamine oxidase with unique features in substrate specificity and N-terminal amino acid sequence. The results strongly suggest that A. terreus NUK-1204 is capable of producing a novel glucosamine oxidase, most suitable for converting glucosamine into glucosaminic acid. Due to its high reactivity and specificity towards glucosamine, the enzyme can also be applied in glucosamine quantification. In addition, given its resistance to H2O2, it can be useful in glucosaminic acid production. Therefore, glucosamine oxidase has high potential for application in pharmaceutical and food production in the future.
Author Statements
The present study was supported by the Ministry of Science and Technology, Taiwan, ROC under Grant no. MOST 110-2320-B-390-001.
References
- Que-Gewirth NLS, Karbarz MJ, Kalb SR, Cotter RJ, Raetz CR. Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A: expression cloning of the outer membrane oxidase LpxQ. J Biol Chem. 2003; 278: 12120-9.
- Zhang JL, Xia WS, Liu P, Cheng QY, Tahirou TB, Gu WX, et al. Chitosan modification and pharmaceutical/biomedical applications. Mar Drugs. 2010; 8: 1962-87.
- Varela O, Nin AP, de Lederkremer RM. A short synthesis of (2S,4S,5R)-4,5,6-trihydroxynorleucine. Tetrahedron Lett. 1994; 35: 9359-62.
- Gerspacher M, Rapoport H. 2-Amino-2-deoxyhexoses as chiral educts for hydroxylated indolizidines. Synthesis of (+)-castanospermine and (+)-6-epicastanospermine. J Org Chem. 1991; 56: 3700-6.
- Burton SG. Oxidizing enzymes as biocatalysts. Trends Biotechnol. 2003; 21: 543-9.
- May SW. Applications of oxidoreductases. Curr Opin Biotechnol. 1999; 10: 370-5.
- Hilt W, Pfleiderer G, Fortnagel P. Glucose dehydrogenase from Bacillus subtilis expressed in Escherichia coli I: Purification, characterization and comparison with glucose dehydrogenase from Bacillus megaterium. Biochim Biophys Acta. 1991; 1076: 298-304.
- Shin KS, Youn HD, Han YH, Kang SO, Hah YC. Purification and characterisation of D-glucose oxidase from white-rot fungus Pleurotus ostreatus. Eur J Biochem. 1993; 215: 747-52.
- Leitner C, Volc J, Haltrich D. Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl Environ Microbiol. 2001; 67: 3636-44.
- Groen BW, De Vries S, Duine JA. Characterization of hexose oxidase from the red seaweed Chondrus crispus. Eur J Biochem. 1997; 244: 858-61.
- Hansen OC, Stougaard P. Hexose oxidase from the red alga Chondrus crispus. Purification, molecular cloning, and expression in Pichia pastoris. J Biol Chem. 1997; 272: 11581-7.
- Horiuchi T. Purification and properties of N-acyl-D-hexosamine oxidase from Pseudomonas sp. 15-1. Agric Biol Chem. 1989; 53: 361-8.
- Danneel H, Ullrich M, Giffhorn F. Goal-oriented screening method for carbohydrate oxidases produced by filamentous fungi. Enzyme Microb Technol. 1992; 14: 898-903.
- Lin SF, Li CK, Chung YP. Identification of a novel lactose oxidase in Myrmecridium flexuosum NUK-21. FEBS Open Bio. 2019; 9: 364-73.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680-5.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951; 193: 265-75.
- Mudgett RE. Solid-state fermentation. In: Demain AL, Solomon NA, editors. Manual of industrial microbiology and biotechnology. Washington, DC: American Society for Microbiology Press. 1986; 66-83.
- Lin SF, Yang TY, Inukai T, Yamasaki M, Tsai YC. Purification and characterization of a novel glucooligosaccharide oxidase from Acremonium strictum T1. Biochim Biophys Acta. 1991; 1118: 41-7.
- Heuts DP, Janssen DB, Fraaije MW. Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by model-inspired site-directed mutagenesis. FEBS Lett. 2007; 581: 4905-9.
- Swoboda BE, Massey V. Purification and properties of the glucose oxidase from Aspergillus niger. J Biol Chem. 1965; 240: 2209-15.