
Research Article
J Bacteriol Mycol. 2019; 6(2): 1101.
Staphylococcus epidermidis Persister Cell Elimination Using an Antimicrobial Peptide
Pihl M and Andersson M*
Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Sweden
*Corresponding author: Martin Andersson, Department of Chemistry and Chemical Engineering, Chalmers University of Technology, Kemivägen 10, SE- 412 96 Göteborg, Sweden
Received: February 13, 2019; Accepted: March 28, 2019; Published: April 04, 2019
Abstract
Biomaterial associated infections are often caused by Staphylococcus epidermidis, which is an opportunistic pathogen with the ability to form biofilms. Such biofilms are inherently resistant to antimicrobials, depending on a variety of factors, including presence of persister cells. Persister cells are dormant, non or slow growing, bacteria that survive long exposures of antibiotics. As an alternative to antibiotics, persisters may be killed using antimicrobial peptides (AMPs). The AMP RRP9W4N was evaluated with the objective to kill both planktonic and biofilm S. epidermidis persister and normal cells. Agar plating and colony counts in addition to fluorescence microscopy were used to determine bacterial survival after exposure to 0-200 μM of the AMP. Scanning electron microscopy was used for morphological bacterial studies. It was demonstrated that 200 μM of the antimicrobial peptide resulted in total planktonic persister elimination and collapse of biofilms, with swelling and bursting of bacteria. Interestingly, persisters seemed more susceptible towards the AMP compared to normal bacteria, another factor underlining the potential use of antimicrobial peptides in future clinical applications.
Keywords: Planktonic bacteria; Biofilm; Dormant; Ciprofloxacin; RP62A
Introduction
The risk of receiving an orthopaedic device-related infection is around 2% [1] and it is an increasing health care problem. In 2009, 566 million USD were spent in the US alone to treat 22’000 infected hip and knee implants, figures that are estimated to reach a cost of 1.62 billion USD in 2020 due to an aging population [2]. One of the most common causes of biomaterial associated infections is the human commensal bacterium Staphylococcus epidermidis and its major pathogenicity factor is related to its ability to form biofilms [3]. Finding new ways to eradicate this opportunistic pathogen is thus of vast importance.
Biofilm formation, extracellular polymer encapsulated bacterial growth on a surface, is causing 80% of all infections [4] and is of great concern when it comes to infection control. Biofilms are inherently resistant to antimicrobials and can be up to 1000 times more resistant to antibiotics compared to their corresponding planktonic counterparts. Biofilm resistance may be caused by different factors such as restricted penetration of the drug through the biofilm, antimicrobial destroying enzymes, efflux pumps, slow growth rate and presence of persister cells [5]. Persister cells are dormant, slow or non-growing, bacteria, which survive prolonged antibiotic treatments. Persisters enriched from a normal population using antibiotics produce a population indistinguishable from the original one with same antibiotic susceptibility, upon regrowth in fresh medium [6]. This indicates persistence is a phenotypic tolerance and not a genetic resistance. Persisters are likely responsible for recurrent biofilm associated infections [7] and antibiotics are usually not efficient in eliminating persisters as they generally act on dividing cells. Hence, new methods to eliminate bacteria, regardless of their metabolic activity, are needed. One possibility is the use of antimicrobial peptides, AMPs. By cleaving peptides from the protein PRELP (proline arginine-rich end leucine-rich repeat protein) and attaching a hydrophobic tail of tryptophan a proteolytically stable peptide with low toxicity for human cells and high bactericidal effect has been obtained, RRP9W4N [8]. This peptide it is active even against multiresistant bacterial strains of Staphylococcus aureus, Group A streptococci, Escherichia coli and Pseudomonas aeruginosa [8].
In this study, we enriched Staphylococcus epidermidis persister cells, both in planktonic and biofilm cultures. Then we investigated the ability of eliminating persisters using the antimicrobial peptide RRP9W4N, using agar plating with bacterial counts and microscopy with LIVE/DEAD staining. Bacterial morphology was investigated using scanning electron microscopy.
Material and Methods
Determination of minimum inhibitory concentration
Staphylococcus epidermidis ATCC 35984 (also known as RP62A) was cultured on Brain Heart Infusion (BHI) agar, 37°C overnight, and then 1 colony was cultured overnight (37°C) in Tryptic Soy Broth (TSB) until mid exponential phase and harvested by centrifugation (2500 rpm, 10 min). The formed pellet was resuspended in fresh TSB and distributed to serially diluted Ciprofloxacin, with final concentrations ranging from 0.03 mg/L to 8 mg/L and a final bacterial concentration of 5*105 CFU/ml. Bacteria were cultured for 24 hours (37°C) before visually inspected for bacterial growth, to determine the minimum inhibitory concentration, MIC. Three independent experiments were performed.
Planktonic persister enrichment
To investigate the proportion of persister cells, one colony of S. epidermidis 35984, obtained from BHI agar, was cultured overnight (37°C) in TSB until stationary phase, harvested by centrifugation (2500 rpm, 10 min) and resuspended in fresh TSB. In total volumes of 4 ml, antibiotic and bacteria were mixed to final concentrations of 2*105 CFU/ml and 0.25, 2.5, 5 and 25 mg/L Ciprofloxacin. Bacteria were cultured for 24 h, 37°C, before serially diluted and cultured on BHI agar plates, to determine CFU/ml. Three independent experiments were performed.
Planktonic bacterial elimination using an antimicrobial peptide
One colony of normal planktonic S. epidermidis 35984, obtained from BHI agar, was cultured overnight (37°C) in TSB until stationary phase and then harvested by centrifugation (2500 rpm, 10 min) and resuspended in fresh TSB. For persister cell enrichment, planktonic bacteria were cultured as above and then exposed to 5 mg/L Ciprofloxacin for 24 h, 37°C, before centrifugation (2500 rpm, 10 min) and resuspension in fresh TSB.
To investigate the antimicrobial effect on normal contra persister cells, they were subjected to an increasing concentration of the antimicrobial peptide RRPRPRPRPWWWW-NH2, RRP9W4N, (Bio-Peptide Ltd). Bacteria were subjected to 0, 25, 50, 100 or 200 μM AMP and cultured for 2 or 24 h (37°C) before serially diluted and cultured on agar plates.
Three independent experiments were made. Statistical analysis was performed using a one-way ANOVA with a Bonferroni post test and P value <0.05 was considered significant.
Biofilm persister enrichment
For biofilm persister cell enrichment, normal bacteria, which were produced as for planktonic bacterial elimination using an AMP, were cultured for 3 h in 8-well glass bottom μ-slides (Ibidi) and then the bacterial solutions were discarded. Biofilms were subjected to fresh medium containing 0, 0.25, 2.5, 5 or 25 mg/L Ciprofloxacin for 24 hours, 37°C, before washed twice in PBS and stained by LIVE/DEAD BacLight (Thermo Fisher Scientific) and studied in a fluorescence widefield microscope (Zeiss). Area covered by dead (red) or live (green) bacteria were evaluated using ImageJ [9]. Three independent experiments were performed.
AMP elimination of biofilm bacteria
Bacteria were cultured as for planktonic bacterial elimination using the AMP. Normal bacteria were then cultured for 3h in 8-well glass bottom μ-slides (Ibidi). For persister enriched bacteria, biofilms were subjected to fresh medium containing 0, 0.25, 2.5, 5 or 25 mg/L Ciprofloxacin for 24 hours, 37°C. The bacterial solutions were then discarded and fresh media containing 0, 25, 50, 100 or 200 μM of the AMP was added to each well.
Both normal and persister enriched biofilms were cultured for 2 or 24 hours (37°C) before washed twice in PBS and stained using LIVE/DEAD BacLight according to the manufacturer (Thermo Fisher Scientific). Viability was studied using a wide field fluorescence microscope (Zeiss). Three independent experiments were performed.
Morphology studies
Normal as well as persister biofilms exposed to 0, 25, 50, 100 or 200 μM AMP were produced as described above and then fixed in 4% buffered formaldehyde overnight in room temperature. The samples were rinsed three times in PBS and subjected to a drying gradient, 50, 60, 70, 80 90 and 100% ethanol for ten minutes per step. Then samples were subjected to 50% hexamethylsilazane in ethanol for 20 minutes and left in 100% hexamethylsilazane until dry after evaporation in ambient air. Samples were gold sputtered for 60s, 10 mA, and studied in SEM (Leo Ultra 55).
Results
MIC determination and planktonic persister formation
Minimum inhibitory concentration of Ciprofloxacin for S. epidermidis 35984 was determined to 0.25 mg/L by visual inspection. This concentration gave the first clear test tube in the antibiotic gradient tested, and all antibiotic concentrations lower than this produced visual bacterial growth.
Stationary phase bacteria are known to produce the most persister cells [10]. When planktonic stationary phase bacteria were exposed to Ciprofloxacin in concentration of the MIC value (0.25 mg/L), 10, 20 and 100 times the MIC, and cultured on agar plates, a small but significant bacterial population was noted, as seen in Figure 1. Only persister cells survive these high antibiotic concentrations [11]. When using 5 mg/L Ciprofloxacin, i.e. 20 times the MIC value, there was an average of 1200 ± 300 CFU/ml left when plated, 0.02% of the total population or a 4-log reduction.
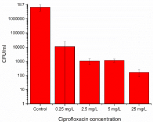
Figure 1: Planktonic bacterial elimination using different concentrations
(MIC=0.25 mg/ml, 10*MIC, 20*MIC and 100*MIC) of Ciprofloxacin for
24 hours revealed presence of a small number of surviving bacteria, the
persister cells.
Planktonic persister AMP elimination
The elimination effect the AMP had on planktonic bacteria was studied by exposure of both normal and persister cells for 2 or 24 hours before serial dilution and agar plating. According to Figure 2, normal planktonic bacteria exposed to the AMP for 2 hours showed a concentration dependent elimination, with a total bacterial extinction at 200 μM AMP. However, already at 25 μM AMP, 98.6% of the normal planktonic cells, were dead. In the normal 24 h control, the number of cells increased compared to the 2 hour control, indicating bacteria being healthy and dividing. This was also true for the lower AMP concentrations of 25 and 50 μM, where the numbers equaled that in the control. These AMP concentrations were not high enough to eliminate the bacteria, which recovered and started to divide. A bacterial reduction was not visible until an AMP concentration of 100 μM, where 99.99% of the cells were eliminated. In 200 μM AMP all 24 h normal cells had been killed and no growth was observed. Furthermore, as seen in Figure 2, for the 2h treated persister cells there is a small downward trend in the number of remaining bacteria, with increasing AMP concentration, until a total elimination at 200 μM.
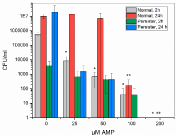
Figure 2: Normal or persister planktonic bacteria were exposed to the AMP,
RRP9W4N, for 2 or 24 hours before cultured on agar to determine CFU/ml.
All bacteria, both normal and persisters, were killed at an AMP concentration
of 200 μM, and all the 24 h persisters were even killed at 100 μM AMP.
*Significant difference to Normal 2h, control. p<0.05. **Significant difference
to Normal 24 h, control. p<0.05.
When comparing the 2 and 24-hour persister controls, with no AMP addition, the bacterial level increased 4 logs after 24 hour growth. However, when exposed to 25 μM AMP the number of persister cells of both 2 and 24 hours were equal. Upon increased AMP concentration there is a downward trend for 24 hour persisters until total extinction after 100 μM. Interestingly, at 100 μM AMP there were still a few surviving 2 hour treated persisters as well as normal bacteria.
Biofilm persister cells were enriched on glass substrates, using Ciprofloxacin. The total area of the Ciprofloxacin exposed biofilms were similar regardless of antibiotic concentration, as seen in Figure 3, and they were constituting 17-33% of the control area. The 20 times MIC value (5 mg/L) was used for continued biofilm persister experiments.
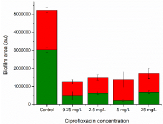
Figure 3: Biofilms were exposed to varying concentrations (MIC, 10*MIC,
20*MIC and 100*MIC) of Ciprofloxacin for 24 hours to enrich the biofilms in
persisters.
Biofilm AMP elimination
When normal biofilm bacteria were exposed to AMP for 2 or 24 hours there was a gradual increase in dead bacteria with increasing AMP concentration, as seen in Figure 4. Compared to the 2 or 24 h persister biofilms, there were more biofilm present in the normal controls, showing that only a low number of bacteria survived the heavy antibiotic treatment to enhance persisters. Interestingly, as seen both for normal and persister biofilms, with increasing AMP concentration irregular, bigger patches were observed in the images. These patches were increasing in number and area with increasing AMP concentration. The persister bacteria produced bigger patches than the normal bacteria, at the corresponding AMP concentration. The size of the patches indicate they were not individual bacteria but consisted of several merged or damaged cells. Due to the patches, no statistics of the biofilm area could be obtained, but only representative images are presented.

Figure 4: Normal and persister enriched biofilms were exposed to increasing concentrations of the AMP. The normal biofilms were more viable at lower AMP
concentrations compared to the persister biofilms. With higher AMP concentration more and larger patches were visible in both normal and persister biofilms.
Images are representative images from three independent experiments. Scale bar is 20 μm.
Morphology studies
The interesting patches seen in the fluorescence microscope entailed morphology studies using SEM. As can be seen in Figure 5, the 2 h normal controls were healthy, spherical cocci. When exposed to the AMP, already at 25 μM, some bacteria started to swell and increase in size, until they finally burst and disintegrated. The number of swelling and disintegrating cells increased with increasing concentration of AMP, although some cells of normal sizes were still found in all samples.

Figure 5: SEM images of bacteria exposed to the AMP RRP9W4N revealed swelling and bursting of bacteria with increasing AMP concentration. Scale bar is 200
nm.
As seen in the fluorescence microscope, the SEM images also revealed the persister cells being affected by the AMP at lower concentrations compared to the normal bacteria. Persisters were also less spherical than normal bacteria. This may be due to the fact that they were already affected by a 24 hour pre-treatment with the antibiotic Ciprofloxacin, and hence were in a more delicate state than the normal bacteria.
Discussion
Persister generation has been extensively studied in E.coli but to a lesser extent in staphylococci [6,12,13,10]. However, since the majority of biomaterial associated infections are related to staphylococci [14] and persisters have been linked to recurrent infections [13], it is of vital importance to investigate persistence of staphylococci. Here, the persistence of S. epidermidis RP62a and the ability of the antimicrobial peptide RRP9W4N to eliminate them was investigated.
Many AMPs are not appropriate for clinical applications due to their cytotoxicity, low bactericidal potency (high MICs), in vivo inactivation by proteases and high production costs [15]. However, end tagging of the peptides with the bulky, aromatic amino acids tryptophan or phenylalanine, prevent the peptides to bind to eukaryotic cholesterol-containing membranes, resulting in decreased cytotoxicity [16]. Hydrophobic amino acid end tagging improves bactericidal potency and by varying the end-tag length proteolytic stability can be achieved [8]. The antimicrobial peptide used in this study, RRP9W4N, was chosen due to its relatively good bactericidal potency, proteolytic stability and low cytotoxicity [8], all factors important for the potential of future clinical use.
The fraction of persisters in a bacterial population is often 10-6- 10-5, but may reach up to 1% in stationary phase cultures for several bacterial species [13,17]. In this study, the fraction of persisters in the stationary phase reached 0.02%. Here the fluoroquinolone Ciprofloxacin was used and it kills both rapidly and slow-growing bacteria [18]. This indicates the potential of also killing some persister cells and is probably the reason for the lower persister fraction obtained in this study. A lower persistence level of S. epidermidis when exposed to fluoroquinolones compared to antibiotics killing rapidly dividing cells has also been seen elsewhere [10]. Although the potential of different antibiotics to kill persister cells varies, per definition persisters are antibiotic-resistant, and thus fruitless to use for total persister eradication. Because of this antibiotics were not included as control in this study, but only used for enhancing the persisters, and it was washed away before AMP exposure. When comparing the 2 and 24 hour persister controls (Figure 2), the bacterial level increased 4 logs after 24 hours, indicating persister cells moving from their dormant state to a dividing state. However, when exposed to 25 μM AMP the number of persister cells of both 2 and 24 hours were equal. Since stress keep persisters in their dormant state, this indicates that even low concentrations of AMP is stressful enough to maintain persistence and that the persisters in this study remain in persistence until elimination.
At a concentration of 80 μM of a dendrimeric peptide, a nearly 3-log reduction of planktonic E. coli persisters was obtained after 1 hour. This AMP was also effective against biofilm persisters, with just above 99% biofilm elimination in one hour [19]. At 200 μM, another synthetic dendrimeric peptide killed between 69-89% of planktonic persisters after 3.5 hours of treatment [20]. In this study, a total planktonic persister elimination is obtained when treated with 100 μM of AMP for 24 hours, or already after two hours when treated with 200 μM of AMP (Figure 2). Total MRSA persister elimination have been found using synthetic antimicrobial peptides [21], and together with our results this indicate the potential for future clinical treatments of persistent infections using AMPs.
For all AMP concentrations there were slightly less planktonic persisters surviving compared to the normal bacteria, both for 2 and 24 hour bacteria (Figure 2). Thus, it seems like persisters are slightly more sensitive to the AMP than the normal bacteria. This phenomenon was also seen for E. coli persister killing, compared to normal bacterial killing using AMPs [19]. Since membrane integrity is vital for the bacteria, regardless of their metabolic activity, to maintain cellular homeostasis [22], we hypothesize that the increased persister susceptibility to the AMP is due to the stalled response of persisters to repair membrane damages due to their metabolic inactivity.
When studying normal biofilms stained with LIVE/DEAD, the AMP seem to have a concentration-dependent killing effect, with more dead bacteria upon increased AMP concentrations. However, with increasing AMP concentration the images also showed larger and larger patches of bacteria, which made statistical comparison of the images difficult. Upon comparing normal and persister biofilms, the persister biofilms seemed more susceptible to the action of AMP, just like seen in Figure 2 for planktonic normal and persister cells. The intriguing patch formation lead us to high-resolution morphology studies using SEM. As seen in the SEM images, gradual bacterial swelling and bursting was observed with increased AMP concentration, in addition to increased presence of small and shrunken bacteria. SEM also revealed the persisters to seem more susceptible to the AMP action.
The first interaction between AMP and microbe occur via electrostatic interactions between the positively charged AMP and negatively charged groups on the bacterial membranes, e.g. lipopolysaccharides and lipoteichoic acids, and via hydrophobic interactions as the peptides are inserted into the membrane. The membrane is disrupted via membrane thinning and poration or disruption of the barrier function, but the AMP may also translocate to targets within the cells [15]. By compromising the membrane, bacteria will take up water in a hypotonic solution, and this will eventually lead to lysis and the cells will burst. Swelling and bursting is seen in the SEM studies, Figure 5, indicating this mechanism of AMP action is also valid for persister cells. As persisters also seem even more susceptible to the action of AMPs, we believe AMP treatment of persistent infections to be of great potential in future clinical settings.
Conclusion
Total persistence elimination can be achieved using the AMP RRP9W4N, both for planktonic cells and biofilms. Interestingly, persisters seem to have a higher susceptibility to AMPs, compared to normal cells. These results, in addition to previous studies on persister elimination using AMPs, [19-21] demonstrate a promising potential use of AMPs in persister eradication in future clinical applications.
Acknowledgement
This work was supported by the Knut and Alice Wallenberg foundation through their Wallenberg Academy Fellows program.
References
- Joseph RL. Prosthetic Joint Infections: Bane of Orthopedists, Challenge for Infectious Disease Specialists', Clinical Infectious Diseases. 2003; 36: 1157-1161.
- Steven MK, Lau E, Watson H, Schmier KJ, Parvizi J. Economic Burden of Periprosthetic Joint Infection in the United States. The Journal of Arthroplasty. 2012; 27: 61-65e1.
- Otto M. Bacterial sensing of antimicrobial peptides. Contrib Microbiol. 2009a; 16: 136-149.
- "Immunology of Biofilms (R01)." In, edited by Depertment of Health and Human Services, PA-07-2007; 288.
- Lewis K. Riddle of biofilm resistance. Antimicrobial Agents and Chemotherapy. 2001; 45: 999-1007.
- Joseph WB. Treatment of Staphylococcal Infections with Penicillin by Intermittent Sterilisation'. The Lancet. 1944; 244: 497-500.
- Lewis K. Persister cells. Annu Rev Microbiol. 2010; 64: 357-372.
- Martin M, Kasetty G, Pasupuleti M, Alenfall J, Schmidtchen A. Highly Selective End-Tagged Antimicrobial Peptides Derived from PRELP. PLoS One. 2011; 6: e16400.
- Johannes S, Carreras IA, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Meth. 2012; 9: 676-682.
- Shapiro JA, Nguyen VL, Chamberlain NR. Evidence for persisters in Staphylococcus epidermidis RP62a planktonic cultures and biofilms. J Med Microbiol. 2011; 60: 950-960.
- Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001; 183: 6746-6751.
- Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation', Antimicrobial Agents and Chemotherapy. 2013; 57: 1468-1473.
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiology Letters. 2004a; 230: 13-18.
- Otto M. Staphylococcus epidermidis-the 'accidental' pathogen. Nat Rev Microbiol, 2009b; 7: 555-567.
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies', Nat Biotechnol. 2006; 24: 1551-1557.
- Pasupuleti M, Chalupka A, Morgelin A, Schmidtchen A, Malmsten A. Tryptophan end-tagging of antimicrobial peptides for increased potency against Pseudomonas aeruginosa, Biochimica et Biophysica Acta. 2009; 1790: 800-808.
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004b; 186: 8172-8180.
- O'Donnell JA, Gelone SP. Fluoroquinolones, Infectious Disease Clinics of North America. 2000; 14: 489-513.
- Chen X, Zhang M, Zhou C, Kallenbach NR, Ren D. Control of bacterial persister cells by Trp/Arg-containing antimicrobial peptides. Appl Environ Microbiol. 2011; 77: 4878-4885.
- Bahar AA, Liu Z, Totsingan F, Buitrago C, Kallenbach N, Ren D. Synthetic dendrimeric peptide active against biofilm and persister cells of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2015; 99: 8125-8135.
- Mohamed MF, Abdelkhalek A, Seleem MN. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep. 2016; 6: 29707.
- Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011; 9: 62-75.