
Research Article
J Bacteriol Mycol. 2016; 3(2): 1024.
Molecular Investigations of Food-Borne Cladosporium and Fusarium Species from Nigeria
Moore GG¹ and Fapohunda SO²*
¹Southern Regional Research Center, Agricultural Research Service, United States Department of Agriculture, New Orleans, Louisiana, USA
²Department of Biosciences and Biotechnology, Babcock University, Ilishan-Remo, Nigeria
*Corresponding author: Stephen O. Fapohunda, Department of Biosciences and Biotechnology, Babcock University, Ilishan-Remo, Nigeria
Received: March 27, 2016; Accepted: May 06, 2016; Published: May 09, 2016
Abstract
A sampling of contaminated foodstuffs throughout southwest Nigeria yielded three fungalisolates belonging to the genus Fusarium and two belonging to the genus Cladosporium. In this study we subjected these isolates to various molecular investigations. The morphological species identifications were confirmed or refined with BLAST queries for sequences from two genomic regions (translation elongation factor-1 α and ITS). BLAST results uncovered species identification inconsistencies for one Fusarium isolate, SRRC1606, and for both Cladosporium isolates based on the examined loci. Using additional species sequences obtained from Gen Bank, the phylogenetic associations for each genomic region were explored and observed species haplotype associations for the Fusarium sequences; however, this was not the case for the Cladosporium sequences. There was evidence of recombination in both loci for the Fusarium species, but only in the translation elongation factor locus for the Cladosporium sample population. Fusarium coalescent analyses for both loci inferred two lineages, one containing only F. oxysporum sequences and the other containing the remaining species examined. These same analyses for the Cladosporia inferred ancient segregation of one lineage, containing only the outgroup taxa, from a second lineage that exhibited recent divergence events among the other taxa examined. Mycological population dynamics and analyses can achieve a better understanding of interventions to protect consumers from contaminated foods.
Keywords: Fungal contaminants; Genotyping; Phylogenetics; Recombination; Speciation
Introduction
The genus Fusarium includes species noted for the production of secondary metabolites and cause of diseases in both plants and animals [1,2], and Cladosporium species are most often associated with cutaneous and pulmonary infections [3]. They occupy very diverse habitats such as plumbing drains [4], tomato plants [5], cosmetics [6], and hospitals [7,8]. Previous research has emphasised the value of molecular techniques, beyond morphological investigation, as critical components for reliable fungal classification. For example, utilization of a molecular technique on Fusarium, known as PCR-ITS-RFLP, was able to further distinguish between strains within the Fusarium Solani Species Complex (FSSC) [9]. A newly developed species-specific primer, for the conserved regions of 28s-rDNA and the intergenic spacer, to genotype F. oxysporum f. sp.psidii was reported by Mishra and co-workers [10]. New genotypes and races of F. Oxysporum f. sp. vasinfectum were discovered, through comparison with previously described races, using sequences from translation elongation factor, phosphate permease, and beta-tubulin genes [11]. Similar attention has been focused on the use of molecular techniques to differentiate Cladosporium species [12-15]. The identities of two races of C. fulvum were recently confirmed by amplifying and sequencing 580bp of their respective ITS regions [5]. Molecular association of metabolite production with fungal genes has also been investigated such as in the FUM-1 (fumonisin) gene, and both TRI-13 and TRI-7trichothecene genes in F. proliferatum and F.verticillioides, respectively [16]. Also, molecular techniques were used to elucidate the genotypes of nonmycotoxigenic strains of F. proliferatum, F.verticillioides, and the F. graminearum species complex strains [17].
The aim of this preliminary study, not disregarding the sample size, was to analyze genomic loci in previously described Fusarium and Cladosporium isolates, sampled from contaminated foodstuffs, to observe species diversity and to assure the integrity of their taxonomic placements. The molecular analysis was to also designed to enhance in -depth fungal knowledge, which is a critical and strategic input in their control and management
Materials and Methods
Fungal identification
Fungal strains selected for this study, originally sampled from contaminated foods in Nigeria, were identified based on morphological characters as in Fapohunda et al. [18] and all isolates are currently housed in the Southern Regional Research Center (SRRC) fungal collection in New Orleans, Louisiana, USA. Three Fusarium isolates (SRRC1606, SRRC1630 and SRRC1633) were morphologically identified from among the collected samples. Also, two Cladosporium isolates were identified based on morphology (SRRC1616 and SRRC1634). In addition to morphology, BLAST queries of sequences from two genomic regions were performed to verify species identification for each isolate: the Internal Transcribed Spacer (ITS) region and the translation elongation factor-1 α (tef1α). The tef1α locus is considered a good delimiter of species for fungal genera such as Fusarium and Cladosporium [19, 20]. All isolates were PCR-amplified for ITS as in Fapohunda et al. [18]. Amplification of the tef1α locus involved primers reported in Carbone and Kohn [21]. Amplicons for each isolate were purified and sequenced, and the sequences were trimmed, cleaned, and aligned using Sequencher version 5.3 (Gene Codes Corporation). Individual sequences for each isolate/locus were BLAST queried to verify species associations previously determined by morphological observations. In this study we did not test any isolates for the presence of mycotoxins.
Comparative analyses
Sampling was done on less than four isolates per genus, and searches for each locus were performed using the NCBI database and additional species sequences from each genus were included in the respective sequence alignments. Whenever possible, the same accessioned isolate (or at least species) was used in alignments for the genomic loci examined. In addition to the various sequences acquired to build sample populations for each genus and locus, outgroup taxa were selected for each genus. Fusarium chlamydosporum [22] and Cladosporium salinae [23] have been reported as appropriate phylogenetic out group taxa for their respective genera, and; therefore, were chosen for this study. All sequence alignments were exported in nexus format for analyses in SNAP Workbench [24]. The first analysis component involved collapsing the sequences into haplotypes using SNAP: Map [25]. Resulting out files were examined for haplotype associations and evidence of the ancestral sequence. The ancestral sequence is evident when its nucleotide composition is identical to the consensus sequence. Next to be performed was the phylogenetic inference for each locus; additionally, bootstrap values and heuristics (e.g., consistency indices and numbers of most-parsimonious trees) were determined using PAUP* software [26]. Comparative analyses of recombination and coalescence were also performed for each locus and sample population with the recombination events for each locus/species group was performed using RecMin [27]. If evidence of recombination was observed, an Ancestral Recombination Graph (ARG) was inferred using the beagle algorithm with one million simulations [28]. Finally, to explore divergence patterns among the species for each locus, without the interference of recombination, coalescent analyses were performed which only consider mutational differences among isolates [29].
The Gen Bank accession numbers for the Fusarium isolates are KP771785-KP771787 (ITS) and KT950249-KT950251 (tef1α). The accession numbers for the Cladosporium isolates are KP771788- KP771789 (ITS) and KT950252-KT950253 (tef1α).
Results
Molecular confirmation of species
Table 1 lists the sampled Nigerian isolates, the food stuffs from which they were sampled and their BLAST identification results.
SRRC ID
Sample Source
ITS
tef1
SRRC1606
Yam
Fusarium equiseti
Fusarium incarnatum
SRRC1630
Cooked rice
F. oxysporum
F. oxysporum
SRRC1633
Bread
F. incarnatum
F. incarnatum
SRRC1616
Cooked rice
Cladosporium cladosporioides
Cladosporium tenuissimum
SRRC1634
Groundnut
C. cladosporioides
c. tenuissimum
Table 1: Sampled Nigerian isolated with sampling sources and identifications based on two genomic loci.
Based on either genomic loci examined, SRRC1630 and SRRC1633 were identified as F. oxysporum or F. incarnatum, respectively. Isolate SRRC1606 identified as F. equiseti based on ITS, but the tef1α sequence indicated this isolate to be F. incarnatum. BLAST queries of the ITS sequences for SRRC1616 and SRRC1634 resulted in their species identifications being C. cladosporioides, but the tef1α sequences for both isolates resulted in different species identifications (C. tenuissimum). All Nigerian sequences have been deposited in Gen Bank and their accession numbers are listed in Tables 2,3.
Haplotype associations
Sequence comparisons, for the Fusarium species, revealed enough polymorphisms within their respective ITS regions to segregate the sequences into six haplotypes, and evidence of the ancestral ITS sequence affiliated with haplotype H5, comprised of isolate SRRC1633 and the two other F. incarnatum sequences. Haplotype designations for the examined Fusarium loci are listed in Table 2. Isolate SRRC1630 shared a haplotype (H2) with the other two F. oxysporum sequences examined. There was enough diversity to segregate the three F. equiseti sequences, including that of SRRC1606, into individual haplotypes (H3, H4 and H6). The tef1αlocus segregated the Fusarium species into seven haplotypes, and no evidence of the ancestral sequence could be observed for this group of fungi. SRRC1606 and SRRC1633 had identical tef1α sequences (H5) and could not be segregated as individuals. This locus resulted in different haplotype segregations compared to ITS. For example, SRRC1630 and the other F. oxysporum sequences segregated into individual haplotypes (H2, H3 and H4) while the F. equiseti sequences shared a single haplotype (H6). For the Cladosporium sequence dataset, a majority of the ITS sequences were identical, resulting in only two haplotypes (Table 3). Haplotype H1 included only the C. salinae sequence, and the second haplotype included all the remaining sequences examined. No evidence of an ancestral sequence was observed for the ITS. The tef1α locus for the Cladosporium sequences were distinct enough to segregate each of the sequences into its own haplotype for both inference types (Table 3). Evidence was observed of the ancestral sequence within a C. tenuissimum sequence (H6) downloaded from Gen Bank when C. salinae was used as the out group taxa, but without this sequence the inference of the ancestral sequence was lost. A total of five C. tenuissimum sequences, including SRRC1616 (H2) and SRRC1634 (H4), examined for this locus were considered as separate individuals.
Species
GenBank Accession
Haplotypes
Phylogeny and ARGa
Genetreeb
ITS
F. Chlamydosporumc
EU715622e
H1
H1
F. equiseti (SRRC1606)d
KP771785
H3
H4
F. equiseti
EU326202e
H4
H3
F. equiseti
KC254029e
H6
H3
F. incarnatum (SRRC1633)d
KP771786
H5
H1
F. incarnatum
KP453980e
H5
H1
F. incarnatum
KJ572780e
H5
H1
F. oxysporum (SRRC1630)d
KP771787
H2
H2
F. oxysporum
EU326216e
H2
H2
F. oxysporum
HQ651161e
H2
H2
tef1α
F. chlamydosporumc
HM134858e
H1
H1
F. equiseti
KR071777e
H6
H5
F. equiseti
KP450714e
H6
H5
F. equiseti
JQ429377e
H6
H5
F. Incarnatum (SRRC1606)d
KT950249
H5
H6
F. incarnatum (SRRC1633)d
KT950250
H5
H6
F. incarnatum
KF255493e
H7
H7
F. oxysporum (SRRC1630)d
KT950251
H2
H2
F. oxysporum
FJ904872e
H4
H4
F. oxysporum
KF728241e
H3
H3
aHaplotypes based on collapsing sequences with not have an inferred phylogeny or ARG.
Loci with fewer than four haplotypes will not have an inferred phylogeny or ARG.
bHaplotypes based on collapsing sequences with excluded indels and excluded infinite sites violations. Loci with fewer than four haplotypes will not have an inferred phylogeny or ARG.
cOut-group species used for each alignment.
dSequences obtained from Nigerian isolates and accessioned in GenBank.
eSequences borrowed from previously accessioned data.
Table 2: Fusarium species and haplotype designations for phylogenies, ARGs and genetrees.
Phylogenetic inference
Using a population framework, phylogenetic inference for the Fusaria at the tef1α locus (Figure 1, Table 2) resulted in the formation of three lineages: F. oxysporum, which included SRRC1630, exhibited greater phylogenetic distance from the other species examined; F. incarnatum, which included SRRC1606 and SRRC1633, shared a clade with the F. equisetisequences; and the outgroup taxa F. chlamydosporum. Strong bootstrap support was observed for each clade. Similar associations were inferred for the ITS phylogeny (Figure 1, Table 2) although clade groupings were not as clear and no bootstrap support could be determined. The tef1α locus for the Cladosporium sequences failed to infer a phylogeny with recoded indels, so two different phylip alignments were created that allowed for phylogenetic inference. For the first, we excluded indels and infinite sites violations in the alignment when collapsing sequences in SNAP: Map, which allowed a phylogeny to be inferred using PAUP*. For the second, we removed C. salinae as the outgroup taxa and instead used a C. cladosporioides sequence downloaded from Gen Bank as the root; we were then able to recode indels during our collapse of the sequences, and subsequently use PAUP* to infer a phylogeny. Both phylogenies are displayed in Figure 2, and their haplotype designations are shown in Table 3. Two Nigerian C. tenuissimum isolates (SRRC1616 and SRRC1634) shared strong cladal associations (H2 and H4, respectively) and bootstrap support, for the tef1α locus (Figure 2A), within a larger clade that included two other C. tenuissimum sequences (H5 and H6) obtained from Gen Bank. However, a fifth C. tenuissimum sequence (H10) shared greater cladal association with two C. cladosporioides sequences (H7 and H9) and marginal association with the C. oxysporum sequence (H8). One C. cladosporioides sequence (H3) and the outgroup taxa (C. salinae) were observed as distinct lineages from the remaining sequences. In Figure 2B we observed similar haplotype associations. For example, the C. Cladosporioides sequence representing H3 in Figure 2A remained a distinct lineage as H1 in (Figure 2B), and the same C. tenuissimum sequence (H10) that shared a cladal association with the other two C. cladosporioides sequences (H7 and H9) in Figure 2A again exhibited close phylogenetic associations in Figure 2B (H7-H9). The differences we observed related to C. oxysporum as a distinct lineage (H6) instead of sharing a clade with other species, and the two Nigerian isolates (C. tenuissimum) exhibited different haplotype groupings instead of grouping together as shown in (Figure 2A). The Cladosporium ITS sequences were too similar and resulted in only two haplotypes for the ten sequences (representing four species); therefore, phylogenetic inference could not be obtained for this region.
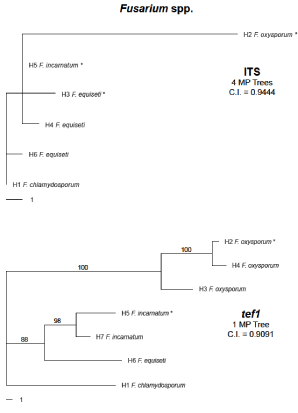
Figure 1: Phylogenies for two genomic regions in multiple Fusarium
species. The outgroup species (H1) used for the inferred trees was F.
chlamydosporum. Mutational distance separating each haplotype can be
determined using the scale bar beneath each tree. Bootstrap values above
70 are displayed at the nodes. Also displayed beneath each locus name is the
Consistency Index (C.I.) value and number of Most Parsimonious (MP) trees
obtained. The haplotype designations and GenBank accessions for each
locus are shown in Table 2.
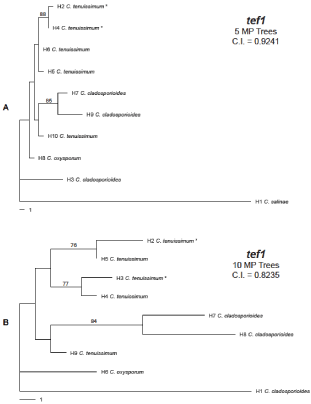
Figure 2: Phylogenies for the tef1α locus in multiple Cladosporium species.
The respective outgroup species (H1) used for inferred tree A was C. salinae
and for inferred tree B was C. cladosporioides. No ITS phylogeny could be
inferred because too few haplotypes resulted when the sequences were
collapsed. Mutational distance separating each haplotype can be determined
using the scale bar beneath each tree. Bootstrap values above 70 are
displayed at the nodes. Also displayed beneath each locus name is the
Consistency Index (C.I.) value and number of Most Parsimonious (MP) trees
obtained. The haplotype designations and GenBank accessions for each tree
are shown in Table 3.
Species
GenBank
Accession
Haplotypes
Phylogeny and ARGa
Genetreeb
ITS
C. salinaec
NR_119606e
H1
H1
C.cladosporioides (SRRC1616)d
KP771788
H2
H2
C. cladosporioides (SRRC1634)d
KP771789
H2
H2
C. cladosporioides
KJ558398e
H2
H2
C. cladosporioides
KF444172e
H2
H2
C. oxysporum
HM148119e
H2
H2
C. tenuissimum
KP131832e
H2
H2
C. tenuissimum
AY545639e
H2
H2
C. tenuissimum
JN033474e
H2
H2
C. tenuissimum
JQ246357e
H2
H2
tef1α (with C. salinae)
C. salinaec
JN906993e
H1b
H1
C. cladosporioides
JF499872e
H3b
H3
C. cladosporioides
HM148290e
H7b
H7
C. cladosporioides
HM148248e
H9b
H9
C. oxysporum
HM148363e
H8b
H8
C. tenuissimum (SRRC1616)d
KT950252
H2b
H2
C. tenuissimum (SRRC1634)d
KT905253
H4b
H4
C. tenuissimum
HM148466e
H10b
H10
C. tenuissimum
HM148461e
H6b
H6
C. tenuissimum
HM148459e
H5b
H5
tef1α (without C. salinae)
C. cladosporioidesc
JF499872e
H1
H1
C. cladosporioides
HM148290e
H7
H7
C. cladosporioides
HM148248e
H8
H8
C. oxysporum
HM148363e
H6
H6
C. tenuissimum (SRRC1616)d
KT950252
H2
H2
C. tenuissimum (SRRC1634)d
KT950253
H3
H3
C. tenuissimum
HM148466e
H9
H9
C. tenuissimum
HM148461e
H5
H5
C. tenuissimum
HM148459e
H4
H4
Table 3: Cladosporium species and haplotype designations for phylogenies, ARGs and genetrees.
Comparative analyses
RecMin analyses among the Fusarium species indicated a minimum of11 recombination events for the tef1α locus, and one for the ITS region; therefore, an ARG was inferred for each (Figure 3, Table 2).
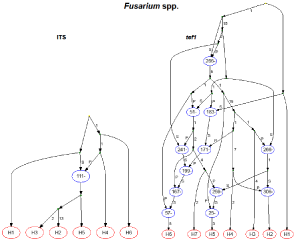
Figure 3: Ancestral Recombination Graphs (ARGs) for two genomic regions
in multiple Fusarium species. Each ARG was inferred using the Beagle
heuristic method for all polymorphisms within the analyzed sequences.
Each ARG shows the possible mutation and recombination paths that
result in the sampled haplotypes (red ovals). The top of the ARG represents
the past and the bottom represents the present. The paths leading to the
recombination nodes (blue ovals) are labelled with a P (prefix) or S (suffix),
indicating the 5’ to 3’ segments of the recombinant sequence, respectively;
the number in each oval indicates the variable position immediately to the
left of the recombination breakpoint. Numbers along the path lines indicate
polymorphisms. The haplotype designations for each locus are shown in
Table 2.
According to the ARG for the tef1α locus, 12 recombination events existed throughout the histories of all species/haplotypes examined, with the exception of the outgroup taxa (F. chlamydosporum, H1). However, evidence existed to suggest the shuffling of the tef1α sequence from F. chlamydosporum into other species/haplotypes. Haplotype H6 (F. equiseti) appeared to result from successive recombination events; and this haplotype, as well as H5 (F. incarnatum), appeared to result from recombination events that occurred on a recent time scale. Haplotype H5 included Nigerian isolates SRRC1606 and SRRC1633. Two F. oxysporum sequences (H3 and H4) experienced a single, ancient recombination event in their respective histories, but the sequence obtained from isolate SRRC1630 resulted from two recombination events. The ARG inferred for the ITS region suggested that haplotypes H1, H4 and H6 (F. chlamydosporum and two F. equiseti Gen Bank sequences, respectively) had no histories of recombination; however, an inferred recombination between H1 and H4 appeared to result in the formation of haplotypes H2 (F. oxysporum which included SRRC1630), H3 (SRRC1606/F. equiseti) and H5 (F. incarnatum which included SRRC1633). RecMin inferred a minimum of eight recombination events for the Cladosporium tef1αlocus that included C. salinae, and its ARG (Figure 4A, Table 3) showed eight recombination events in the histories of haplotypes H3, H7 and H9 (C. cladosporioides), as well as H8 (C. oxysporum), with the most recent recombination events resulting in haplotypes H3 and H8. The remaining haplotypes represented the C. salinae and C. tenuissimum (including SRRC1616 and SRRC1634) species sequences which were not inferred to result from previous recombination events, but they had contributed genetic material to the recombinant sequences mentioned above. RecMin inferred a minimum of six recombination events for the alignment without C. salinae as the outgroup taxa, and the ARG revealed eight possible recombination events (Figure 4B), but the inferences change in regard to most of the haplotypes. Four haplotypes were inferred to have histories of recombination in (Figure 4A), but in (Figure 4B) that number changes to five. The C. cladosporioides isolate represented as H3 in (Figure 4A) was inferred to have derived from a recent recombination event, but this isolate was considered the root for the inferences in (Figure 4B). In these types of analyses it is assumed that the root is not a recombinant. Nigerian isolate SRRC1616 was not inferred to have a history of recombination for (Figure 4A), but was inferred to result from a recent recombination in (Figure 4B). The only two haplotypes that were consistently inferred to have no histories of recombination were H4 and H10 (Figure 4A) and H3 and H9 (Figure 4B). These haplotypes, for both ARGs, contained the same representative sequences (Table 3). Since the Cladosporium ITS region resulted in too few haplotypes, no recombination analyses could be performed.
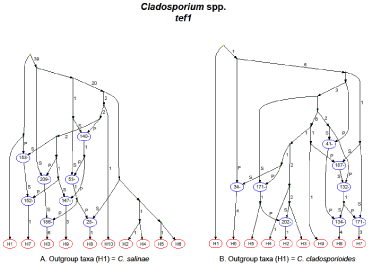
Figure 4: Ancestral recombination graphs for the tef1α locus in multiple
Cladosporium species. The respective outgroup species (H1) used for
inferred ARG A was C. salinae and for inferred ARG B was C. cladosporioides.
No ITS ARGs could be inferred because too few haplotypes resulted when
the sequences were collapsed. The ARGs were inferred using the Beagle
heuristic method for all polymorphisms within the sequences analyzed. Each
ARG shows the possible mutation and recombination paths that result in the
sampled haplotypes (red ovals). The top of the ARG represents the past and
the bottom represents the present. The paths leading to the recombination
nodes (blue ovals) are labeled with a P (prefix) or S (suffix), indicating the 5’ to
3’ segments of the recombinant sequence, respectively; the number in each
oval indicates the variable position immediately to the left of the recombination
breakpoint. Numbers along the path lines indicate polymorphisms. The
haplotype designations for each ARG are shown in Table 3.
In the coalescent analyses and haplotype designations for the Fusarium species as shown in Figure 5, Table 2. Two Fusarium lineages for the tef1α region were observed, with F. oxysporum being the sole species represented in one of them, while the second lineage experienced a series of speciation events that resulted in the other Fusaria examined. The F. oxysporum lineage had experienced bifurcations that segregated into three haplotypes (H2-H4). The other lineage showed the evolutionary time point when F. chlamydosporum (H1) diverged from F. equiseti (H5) as well as the time point when F. equiseti diverged from what is now F. incarnatum. Even F. incarnatum exhibited a bifurcation into two distinct branches represented by haplotypes H6 (Nigerian isolates) and H7. The ITS locus also inferred two ancient lineages whereby F. oxysporum (H2), as a species/complex, had persisted as its own lineage. The second lineage offered little resolution for the remaining species examined, because a single bifurcation, occurring millions of years ago, gave rise to three branches undergoing a clonal amplification. Two of those branches are represented by F. equiseti (H3 and H4/SRRC1606) and the third branch showed sequence identity for F. chlamydosporum and F. Incarnatum (H1). The Cladosporium tef1αgenetree for which C. salinae was the outgroup taxa (Figure 6A, Table 3) inferred C. salinae as an ancient species and sole representative of its own lineage, while the other species shared a second lineage.
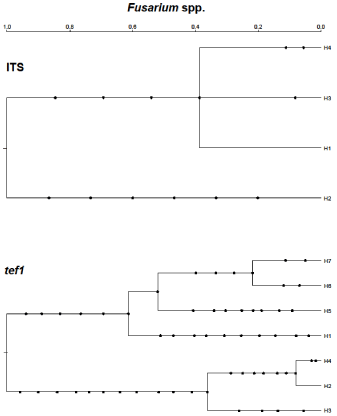
Figure 5: Gene genealogies for two genomic regions in multiple Fusarium
species. These coalescent-based trees show mutations as dots along
branches and time is scaled to The Most Recent Common Ancestor (TMRCA)
of 1.0 for each locus. The haplotype designations for each locus are shown
in Table 2.
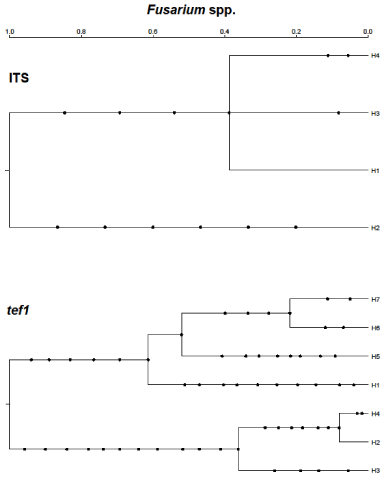
Figure 6: Gene genealogies for the tef1α locusin multiple Cladosporium
species. The respective outgroup species (H1) used for inferred gene tree
A was C. salinae and for inferred gene tree B was C. cladosporioides. No
ITS ARGs could be inferred because too few haplotypes resulted when the
sequences were collapsed. These coalescent-based trees show mutations
as dots along branches and time is scaled to The Most Recent Common
Ancestor (TMRCA) of 1.0 for each locus. The haplotype designations for each
locus are shown in Table 3.
A fairly recent bifurcation in the second lineage appeared to segregate C. tenuissimum into multiple haplotype branches. One of those branches showed the speciation events associated with C. cladosporioides and C. oxysporum. The most recent bifurcation for this group resulted in the segregation of the Nigerian isolates (H2 and H4).The gene tree without C. salinae (Figure 6B) offered more resolution for the group of fungi sharing the second lineage in Figure 6A. We observed bifurcations resulting in haplotypes H1-H6. Haplotypes H7-H9 were not resolved and appeared to undergo clonal amplification since their divergence from H2-H5. No gene tree could be inferred for the ITS region due to having only two haplotypes.
Discussion
It has been shown, through these investigations, that two of the Nigerian Fusarium isolates were consistently identified using different genomic loci. This would indicate that ITS and tef1α are adequate species delimiters for this particular genus. The exception for this group of isolates was SRRC1606, identified as F. equiseti based on ITS sequence, but also as F. incarnatum based on its tef1αsequence. Both species are part of the Fusarium Incarnatum-Equiseti Species Complex (FIESC) [30] and share a most recent common ancestor. In SRRC1606 we see evidence of recombination whereby the ancestry of the ITS region originated with an F. equiseti strain and the tef1α locus originated with an F. incarnatum strain. Reports of F. equiseti and F. incarnatum as specifically pathogenic to yams are lacking although both species have been reported as pathogens of potatoes [31,32]. Some of the mycotoxins associated with F. equiseti are nivalenol, A and B trichothecenes and zearalenone [31,33], while F. incarnatum has been reported to produce beauvericin and zearalenone [32]. Isolate SRRC1633 was sampled from bread and identified as F. incarnatum based on ITS and tef1α regions. Although reports of F. incarnatum as a cereal grain pathogen are rare, they do exist [34]. Isolate SRRC1616 was consistently identified as F. oxysporum based on both genomic regions examined. This isolate/species was sampled from cooked rice which correlated with its reported pathogenicity of cereal grains, and beauvericin and moniliformin are among the mycotoxins that may be produced by F. Oxysporum [32,35]. ITS and tef1α did not offer consistency for species delimitation for sampled Cladosporium species from Nigeria. We determined the respective identifications of isolates SRRC1616 and SRRC1634 as C. Cladosporioides (ITS) and C. Tenuissimum (tef1α). Taxonomically, C. Cladosporioides shares no synonymy with C. tenuissimum, which supported that these are distinct species. However, these two species are part of the same (C. cladosporioides) complex [36] and share a most recent common ancestor. Interestingly, no evidence of recombination was observed in the histories of these isolates. This likely relates to the necessity of removing indels and infinite sites violations in order to proceed with our haplotype associations and comparative inferences. SRRC1616 was sampled from cooked rice and SRRC1634 was sampled from groundnut (peanut). Our findings supported previous reports of a host-pathogen relationship between these two fungal species and cereal grains such as rice, but only C. cladosporioides has been reported on groundnut [37,38]. There have been no reported mycotoxins produced by Cladosporium species [39]. The exact reason for two conserved loci offering different taxonomy is uncertain, but may be the results of ancient inter specific recombination, or horizontal gene transfer, of the loci examined from one species to another that has maintained in isolates like SRRC1606, SRRC1616 and SRRC1634.
From a phylogenetic perspective, the Fusaria exhibited an expected segregation for the loci examined. Cladal associations could be observed, and overall the species/complex distinctions were maintained. For example, F. Chlamydosporum Represents Its Own Species Complex (FCSC) but is considered a closely-related lineage to the FIESC complex that includes the F. incarnatum and F. equiseti species [30]. The most divergent lineage of species examined was F. oxysporum which represents its own species complex (FOSC). Inferring phylogenetic association for the Cladosporia; we were unable to infer a phylogeny for the ITS region because 99% of the sequences obtained were identical. The examined species are encompassed within the C. cladosporioides species complex. The observation that not enough haplotype diversity existed for the Cladosporium ITS region suggests that these are indeed closely-related species whose diversity will likely exist in other loci. For example, we observed greater sequence diversity for the tef1α locus. As expected for closelyrelated species, two C. Cladosporioides sequences were observed sharing a clade with C. tenuissimum while a third C. cladosporioides sequence was inferred as its own lineage. The high degree of diversity among C. tenuissimum sequences could relate to geography or host specificity, but could also relate to misidentifications which can occur for closely-related species. Future studies should involve an increase in the numbers of sampled isolates for more refined inferences of intra-species population dynamics.
Contemporary recombination is an occurrence that is more likely within panmictic populations, but with conserved loci it is still possible to infer ancient recombination events, despite geographic isolation, among coevolved species [39,40]. We were able to infer recombination within the histories of multiple Fusarium species based on two different genomic regions that are considered “conserved” [41]. This finding would support that (1) historical recombination likely contributed to species diversity among these fungi [42,43], and (2) true species delimitation for these fungi would require genomescale comparisons and a more holistic approach to identification [44]. Recombination among Cladosporium species was only observed for the tef1α locus, but the richness of species diversity may require examining other loci and additional species. Based on our inferences of recombination, the different species within each genus, are either exhibiting ancient genetic configurations that have been maintained in certain species, or are exchanging genetic material on a recent time scale as part of their convolution. This is because they share a common ancestor,
Coalescent analysis removes the influence of recombination from species evolution by simply counting mutations and applying them to a time scale based on the assumption of a neutral mutation rate [29]. Basically, the greater the number of mutations along a sequence, the longer that sequence has been in existence. The finding that ITS sequences from three F. Oxysporum isolates segregated as a distinct lineage, with no divergence, suggests it is an ancient organism that long ago diverged from the common ancestor of the other examined Fusarium species. Despite being species from closelyrelated complexes, F. chlamydosporum, F. equiseti and F. incarnatum exhibited recent divergence of their respective ITS regions with no further resolution. More evident patterns of divergence/speciation were observed for the tef1α locus. For example, in the F. oxysporum lineage isolate SRRC1630 was derived from a recent divergence event. The divergence of FCSC and FIESC occurred millions of years ago, and not too long after their divergence a speciation event segregated F. equiseti and F. incarnatum. The tef1α locus for the Cladosporia also suggested an ancient divergence for which the C. salinae lineage is shown as much older than the other species examined. Resolution of the divergence of the remaining Cladosporium species is moderate, but what can be inferred are recent divergence/speciation events for species that share a complex. As well, the Nigerian isolates appeared to be the most recently derived.
In conclusion, the sampling of Fusarium species on foodstuffs in Nigeria is import when one considers the potential for the identified species to produce mycotoxins. We did not perform any mycotoxin assays for our sampled isolates in this study. The identification of a fungus on a particular host for which it is not considered a pathogen may indicate the presence of new species that happen to share genomic sequence with a known species/pathogen. Our findings support the need for a more holistic approach to species identification; particularly, when those fungi are contaminating foods and feeds and even cosmetics [45].
Author Contribution
GG Moore and SO Fapohunda conceived and designed the experiment. Fapohunda did the isolation of the fungi from the food samples while Moore performed molecular analyses. Both contributed to the writing of the paper.
Acknowledgements
The authors would like to thank Mrs. Shannon B. Beltz for her assistance in the morphological identification of the Nigerian isolates, and all reviewers who helped to improve this body of work.
References
- Kovalsky Paris MP, Schweiger W, Hametner C, Stückler R, Muehlbauer GJ, Varga E . Zearalenone-16-O-glucoside: a new masked mycotoxin. J Agric Food Chem. 2014; 62: 1181-1189.
- Tamura M, Naoki M, Nagatomi Y, Harayama K, Akira T, Hayakawa K. A method for simultaneous determination of 20 Fusarium toxins in cereals by High-Resolution Liquid Chromatography-Orbitrap Mass Spectrometry with a pentafluorophenyl column. Toxins. 2015; 7: 1664-1682.
- de Hoog GS, Guarro J, Gené J, Figueras MJ. Atlas of Clinical Fungi. 2nd ed. Utrecht, The Netherlands: Centraalbureau voor Schimmel cultures. 2000.
- Short DP, O'Donnell K, Zhang N, Juba JH, Geiser DM. Widespread occurrence of diverse human pathogenic types of the fungus Fusarium detected in plumbing drains. J Clin Microbiol. 2011; 49: 4264-4272.
- Rollan C, Protto V, Medina R, Lopez S, Vera-Bahima J, Ronco L, et al. Identification of races 0 and 2 of Cladosporium fulvum (syn Passalora fulva) on tomato in the Cinturón Hortícola de La Plata, Argentina. Plant Dis. 2013; 97: 992.
- Pinto FCJ, Braga de Lima D, Agustini BC, Dallagassa CB, Shimabukuro MF, Chimelli M, Morphological and molecular identification of filamentous fungi isolated from cosmetic powders. Braz Arch Biol Technol 2012; 55: 897-901.
- Migheli Q, Balmas V, Harak H, Sanna S, Scherm B, Aoki T . Molecular phylogenetic diversity of dermatologic and other human pathogenic fusarial isolates from hospitals in northern and central Italy. J Clin Microbiol. 2010; 48: 1076-1084.
- Oechsler RA, Feilmeier MR, Ledee DR, Miller D, Diaz MR, Fini ME . Utility of molecular sequence analysis of the ITS rRNA region for identification of Fusarium spp. from ocular sources. Invest Ophthalmol Vis Sci. 2009; 50: 2230-2236.
- Chehri K, Salleh B, Yli-Mattila T, Reddy KRN, Abbasi S. Molecular characterization of pathogenic Fusarium species in cucurbit plants from Kermanshah province, Iran. Saudi J Biol Sci. 2011; 18: 341-351.
- Mishra RK, Pandey BK, Singh V, Mathew AJ, Pathak N, Zeeshan M . Molecular detection and genotyping of Fusarium oxysporum f. sp. psidii isolates from different agro-ecological regions of India. J Microbiol. 2013; 51: 405-412.
- Holmes EA, Bennett RS, Spurgeon DW, Colyer PD, Davis RM. Newgeno types of Fusarium oxysporum f. sp. vasinfectum from the southeastern United States. Plant Dis. 2009; 93: 1298-1304.
- Braun U. Cladosporium exoasci, C. exobasidii and some allied species. Schlechtendalia. 2001; 7: 53-58.
- Braun U, Crous PW, Dugan FM, Groenewald JZ, de Hoog GS. Phylogeny and taxonomy of cladosporium-like hyphomycetes, including Davidiella gen Nov, the teleomorph of Cladosporium s.str. Mycol Progr 2003; 2: 3-18.
- Braun U, Schubert K. Taxonomic revision of the genus Cladosporium s. lat. 7. Descriptions of new species, a new combination and further new data. Schlechtendalia. 2007; 16: 61-76.
- Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol. 2010; 67:1-94.
- Sampietro DA, Marín P, Iglesias J, Presello DA, Vattuone MA, Catalán CA, et al. A molecular based strategy for rapid diagnosis of toxigenic Fusarium species associated to cereal grains from Argentina. Fungal Biol. 2010; 114: 74-81.
- Sampietro DA, Ficoseco ME, Jimenez CM, Vattuone MA, Catalán CA. Trichothecene genotypes and chemo types in Fusarium graminearum complex strains isolated from maize fields of northwest Argentina. Int J Food Microbiol. 2012; 153: 229-233.
- Fapohunda SO, Moore GG, Ganiyu OT, Beltz SB. Toxigenic Aspergillus flavus and other fungi of public health concern in food and organic matter in southwest Nigeria. Mycology: An Int J. Fungal Biol. 2012; 3: 210-219.
- Castaño R, Scherm B, Avilés M. Genetic Diversity of Fusarium oxysporum f. sp. dianthi in Southern Spain, J Mycol. 2014.
- Schoch CL, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW . A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia. 2006; 98: 1041-1052.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999; 91: 553-556.
- Darvishnia M. Morphological and Phylogenetic Studies of Fusarium species in Iran. J. Novel Appl Sci 2013; 2: 1134-1142.
- Zalar P, de Hoog GS, Schroers H-J, Crous PW, Groenwald JZ, Gunde-Cimerman M. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol. 2007; 58: 157-183.
- Price EW, Carbone I. SNAP: workbench management tool for evolutionary population genetic analysis. Bioinformatics. 2005; 21: 402-404.
- Aylor DL, Price EW, Carbone I. SNAP: Combine and Map modules for multilocus population genetic analysis. Bioinformatics. 2006; 22: 1399-1401.
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0. 2003; Sinauer Associates, Sunderland, Massachusetts, 2003.
- Myers SR1, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003; 163: 375-394.
- Lyngsø RB, Song YS, Hein J. Minimum recombination histories by branch and bound. Proceedings of the 5th International Workshop on Algorithms in Bioinformatics (Lecture Notes in Bioinformatics 3692). 2005; 239-250.
- Griffiths RC, Tavaré S. Ancestral inference in population genetics. Stat, Sci. 1994; 9: 307-319.
- O'Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F.equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol 2009; 47: 3851-3861.
- Goswami RS, Dong Y, Punja ZK. Host range and mycotoxin production by Fusarium equiseti isolates originating from ginseng fields. Can J Plant Pathol 2008; 30: 155-160.
- Thrane U. Fusarium. In: Batt CA, Tortorello ML, editors. Encyclopedia of Food Microbiology. New York: Academic Press. 2014. 76-81.
- Kristensen R, Torp M, Kosiak B, Holst-Jensen A . Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycol Res. 2005; 109: 173-186.
- Mackinaite R, Kacergius A, Lugauskas A, Repeckiene J. Contamination of cereal grain by Fusarium. Micromycetes and their mycotoxins under Lithuanian climatic conditions. Ekologija. 2006; 3: 71-79.
- Hsuan HM, Zakaria L, Salleh B. Characterization of Fusarium isolates from rice, sugarcane and maize using RFLP-IGS. J Plant Prot Res. 2010; 50: 409-415.
- Sandoval-Denis M, Sutton DA, Martin-Vicente A, Cano-Lira JF, Wiederhold N, Guarro J. Cladosporium Species Recovered from Clinical Samples in the United States. J Clin Microbiol. 2015; 53: 2990-3000.
- Pitt JI, Hocking AD. Fungi and food spoilage. 2nd ed. Sydney: Springer Press; 1997.
- Stone JK, Bacon CW, White, JF. An overview of endophytic microbes: Endophytism defined. In: Bacon CW, White JF, editors: Microbial Endophytes. New York: Marcel Dekker Inc. 2000; 3-29.
- Milgroom MG. Recombination and the multilocus structure of fungal populations. Annu Rev Phytopathol. 1996; 34: 457-477.
- Carbone I, Kohn L. Inferring process from pattern in fungal population genetics. Appl Mycol. Biotechnol 2004; 4: 1-30.
- Bradshaw RE, Foster SJ, Monahan BJ. Molecular diagnostic tools for detection of plant pathogenic fungi. In Rao JR, Fleming CC, Moore JE, editors. Molecular Diagnostics: Current Technology and Applications. Norfolk, United Kingdom: Horizon Bioscience. 2006. 47-70.
- Kerényi Z, Moretti A, Waalwijk C, Oláh B, Hornok L. Mating type sequences in asexually reproducing Fusarium species. Appl Environ Microbiol. 2004; 70: 4419-4423.
- Stergiopoulos I, De Kock MJ, Lindhout P, De Wit PJ. Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol Plant Microbe Interact 2007; 20: 1271-1283.
- Samson RA, Varga J. What is a species in Aspergillus? Med Mycol. 2009; 47 Suppl 1: S13-20.
- Pinto F C, de Lima DB, Agustini BC, Dallagassa CBI, Shimabukuro MF, Chimelli. TM Morphological and molecular identification of filamentous fungi isolated from cosmetic powders. Braz. Arch. Biol. Technol. 2012; 55.