
Review Article
J Bacteriol Mycol. 2025; 12(1): 1225.
Signaling Mechanisms of Bacteria & Fungi: A Comprehensive Overview
Anushka A, Rojina K, Sudeshna S and Malavika B*
Department of Biotechnology, Techno India University, EM-4, Sector-V, Salt Lake, Kolkata, West Bengal-700091, India
*Corresponding author: Malavika Bhattacharya, Department of Biotechnology, Techno India University, EM-4, Sector-V, Salt Lake, Kolkata, West Bengal-700091, India Email: malavikab@gmail.com
Received: June 04, 2025 Accepted: July 13, 2025 Published: July 17, 2025
Abstract
There are several cellular mechanisms referred to as signalling pathways that have been identified in microorganisms, including fungi and bacteria, for controlling various properties such as resistance to antibiotics, pathogenicity, biofilm formation, and morphological differentiation. Biotechnology, medicine, and agriculture hold highly promising applications of such signalingpathways. Quorum sensing (QS) presents a communication network in bacterial species, which senses autoinducers, signaling molecules, allowing for intercellular communication and environmental stimulus response. In this context, bacteria have emerged as an important target for therapy against virulence targets with the hope of increasing their susceptibility to antimicrobial substances. Quorum quenching (QQ) is a strategy of countering bacterial infections by interfering with QS-signaling pathways in populations of bacteria. The other signaling pathway that was involved in signaling across the membrane in bacteria is the Two-Component System (TCS). TCS typically contains sensor histidine kinases responsive to extracellular stimuli, which, after autophosphorylation of response regulators results in the regulation of transcription. Mechanisms of Fungal Signal Transduction Many areas of healthcare and biotechnology are engaging with high interest in understanding the mechanisms of fungal signaling. Protein kinase A/cAMP, protein kinase C/MAPK, and, calcium-calcineurin pathways regulate virulence characteristics among the different species of pathogenic fungi. Identifying these pathways may hold positive benefits in reducing infections by microbes, improving industrial processes, and advancing the practice of agriculture and healthcare. We discuss the complexities of such complex signalling mechanisms, their utility, and their promise for future applications.
Keywords: Quorum sensing; Quorum quenching; Protein kinase A/ cAMP; Protein kinase C/ MAPK
Introduction
Micro-organisms are sometimes referred to as microbes. They involve living organisms that one cannot see with naked eyes but is visible under a microscope [1]. They include bacteria, viru. There are several cellular mechanisms referred to as signaling pathways that have been identified in microorganisms, including fungi and bacteria, for controlling various properties such as resistance to antibiotics, pathogenicity, biofilm formation, and morphological differentiation. Biotechnology, medicine, and agriculture hold highly promising applications of such signaling pathways. Quorum sensing (QS) presents a communication network in bacterial species, which senses autoinducers, signaling molecules, allowing for intercellular communication and environmental stimulus response. In this context, bacteria have emerged as an important target for therapy against virulence targets with the hope of increasing their susceptibility to antimicrobial substances. Quorum quenching (QQ) is a strategy of countering bacterial infections by interfering with QS-signaling pathways in populations of bacteria. The other signaling pathway that was involved in signaling across the membrane in bacteria is the Two-Component System (TCS). TCS typically contains sensor histidine kinases (HKs) responsive to extracellular stimuli, which, after autophosphorylation of response regulators (RRs), results in the regulation of transcription. Mechanisms of Fungal Signal Transduction Many areas of healthcare and biotechnology are engaging with high interest in understanding the mechanisms of fungal signaling. Protein kinase A/cAMP, protein kinase C/MAPK, and, calcium-calcineurin pathways regulate virulence characteristics among the different species of pathogenic fungi. Identifying these pathways may hold positive benefits in reducing infections by microbes, improving industrial processes, and advancing the practice of agriculture and healthcare. In this chapter, we discuss in detail some of the complexities of such complex signalling mechanisms, their utility, and their promise for future applications. Bacteria are microscopic organisms that have long been the focus of scientific research and medical concern because they can infect hosts with compromised immune systems [2]. Fungi, on the other hand, are organisms that can digest food externally and receive nutrients via their cell walls. They can also obtain carbon and energy from different species [3]. All living things, even microorganisms like bacteria and fungi, have intricate cellular systems called signaling mechanisms that allow them to react to stressors and changes in their environment. Some of the chemicals involved in the process include calcium ions, hormones, reactive oxygen species, and nitric oxide [4]. Signaling pathways in bacteria and fungi are distinguished by functional pleiotropy, in which components serve many purposes and can react to different stimuli using common network members [5].
Microbial signaling pathways are essential for controlling different microbial interactions and functions. These processes have a variety of effects on biofilm formation, host-pathogen interactions, and microbial ecology [6,7].
Bacteria employ a mechanism known as quorum sensing, which enables bacteria to communicate with each other thereby regulating gene expression based on population density [8,9]. However, disrupting QS systems synonymously known as quorum quenching (QQ), has emerged as an effective strategy to combat bacterial infections by squashing microbial signaling [10,11,12].
Bacteria also possess two-component signaling systems (TCS) that have regulatory characteristics, like feedback loops and HK bifunctionality, thus producing complex reactions [13].
Similarly, in case of fungi, fungal signaling pathways are essential for pathogenicity, growth, and environmental sensing. Protein kinase A/cAMP, protein kinase C/MAPK, and calciumcalcineurin are the signaling pathways that control virulence characteristics in different kinds of fungi including pathogenic fungi such as Cryptococcus neoformans [14]. The pleiotropic nature of the signaling proteins and the range of responses elicited by a single stimulus further underline the complexity of fungal signaling [5].
The discovery of these pathways might be able to positively impact the mitigation of infections through microbes, enhance industrial activities, and innovation in agriculture and health.
Signaling Mechanisms in Bacteria
Quorum Sensing
In the past, bacterial cells were thought to be independent living things capable of evolving and metabolizing through self-regulatory mechanisms but it is now widely known that bacterial cells use a technique known as quorum sensing to detect and create tiny signaling molecules known as autoinducers [15]. The primary effects of QS signal emission and sensing are that they control the synthesis of a variety of extracellular factors, which are expelled from cells and serve a variety of purposes such as virulence, immunological suppression, scavenging for resources, scaffolding for surfaceassociated microorganisms (biofilms), and promoting motility [16].
Autoinducers of Bacterial Quorum Sensing: Two primary categories of autoinducers have been brought to light: in gram-positive bacteria, we have oligopeptides whereas in gram-negative bacteria, we have AHL which stands for acylated homoserine lactones [8]. Histidine kinase receptors that are membrane-bound or cytoplasmic transcription factors are responsible for detecting autoinducers [17]. AHLs are detected by specific receptors and generated by LuxI-type synthases [18].
An essential component of bacterial quorum sensing is the LuxS enzyme that catalyzes the synthesis of the precursor autoinducer-2(AI-2) [19]. A universal signal molecule, it's a homodimeric protein with an unusual fold based upon an eight-stranded beta barrel, as determined by the crystal structure of LuxS [19,20]. It can be used not only by Gram-positive bacteria but also by Gramnegative bacteria in interspecies communication [21]. The His54, His58, and Cys126 are conserved residues that coordinate the presence of a zinc ion at each active site [19]. Through a sequence of isomerization and elimination events, LuxS catalyzes the cleavage of S-ribosyl homocysteine, with the metal ion functioning as a Lewis acid [22]. Since the structure and function of the enzyme are extensively preserved among bacterial species, it has been considered a potential good target for the creation of antibiotics [19,23].
Two examples of such applications include the fusion and testing of p-coumaroyl homoserine lactone and N-3-oxo oct – 7 – enoyl – L – homoserine lactone for quorum-sensing activity of Vibrio fischeri [24].
Quorum Sensing in Gram-positive Bacteria: Gram positive bacteria comprise a group of bacteria that have thick peptidoglycans; their peptidoglycan retains the crystal violet stain when the bacterium is stained [25]. To showcase proteins on the surface of the cell wall, gram positive bacteria have developed a number of ways, from lipoproteins, LPTXG- like proteins, transmembrane and cell-wall binding proteins [26].
QS, in gram positive bacteria is a cell density dependent process that regulates several physiological processes for instance, the biosynthesis of virulence factors and the production of biofilms [27]. The QS circuit generally includes inducers such as peptide pheromones that work as signaling molecules; a receptor protein that activates the target genes on binding; and two-component signaltransduction systems [27]. These peptide signals are called autoinducing peptides or AIPs. They are processed post-translationally and are released using ATP-binding-cassette exporters. After secretion, these peptides are either taken back by oligopeptide transport systems or sensed by two-component systems on the bacterial surface [28]. Internalized peptides have interactions with cognate regulators [28].
The use of these systems relies on a peptide signal that induces celldensity dependent expression of genes [29,30]. AIPs accumulate in the extracellular space as the populations of the bacteria increase until the maximum concentration levels are achieved as shown in Figure 1 [8]. Membrane-bound histidine kinases are components of twocomponent signal-transduction systems that later sense these peptide signals [31]. Upon binding AIP, the sensor kinase phosphorylates its corresponding response regulator, activates it, and brings about alterations in gene expression [29].
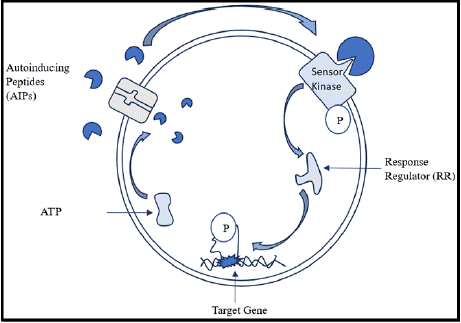
Figure 1: Diagrammatic representation of mechanism of quorum sensing in
gram positive bacteria.
Understanding QS in gram positive bacteria will lead the way towards the development of targeted treatments, opening many doors to applications in industries.
Quorum Sensing in Gram-negative Bacteria
Gram negative bacteria are a type of microbes whose cell walls contain an outer lipoid membrane and a thin layer of peptidoglycan, whereas gram positive bacteria have no outer lipoid membrane with a thick layer of peptidoglycan [32,33]. Gram-negative bacteria's growing resistance to currently existing antibiotics emphasizes the necessity of using antibiotics sparingly and developing new medications [34].
Gram negative bacteria use the QS mechanism as a form of communication in controlling group behaviour based on population density [35]. During the process, it relies on the production and detection of the autoinducers- the most prevalent ones being acyl- HSLs in gram negative bacteria [36,37]. By detecting the concentration of signal molecules present in the surroundings, the mechanism enables bacteria to keep an eye on the growth of their population [37]. This QS system regulates many physiological functions, such as biofilm formation, motility, and synthesis of virulence factors [38,39]. The design and integrity of biofilms are influenced by AHLs, and their absence may make biofilms more vulnerable to antibacterial treatments [38].
Mechanism: LuxI-type enzymes produce AHLs, which build up in the extracellular space as cell density rises as shown in Figure 2 [40]. These ligands are said to bind to intracellular receptors of LuxR-type, which are transcriptional activators when the level of AHL is above threshold [41]. The two functional domains include an N-terminal ligand-binding domain and a C-terminal DNA-binding domain containing a helix-turn-helix motif of the LuxR-type proteins [42]. roteins and leads to either activation or repression of the target genes [40].
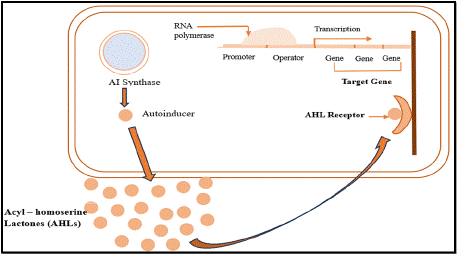
Figure 2: Diagrammatic representation of mechanism of quorum sensing in
gram negative bacteria.
Quorum Quenching
of the promising strategies to control bacterial infection. It works by interfering and inhibiting quorum sensing in bacteria [43]. Signal production, transportation, accumulation, recognition, and autoinduction are just a few of the operations in the QS circuit that QQ can target [43].
The use of QQ techniques has demonstrated potential in a number of domains, such as wastewater treatment, human pathogenic bacterial attenuation, and agricultural and aquaculture pathogen biocontrol [44].
Lactonases, acylases, and oxidoreductases are the three primary classes of QQ enzymes that have been discovered [45,46]. These enzymes efficiently silence QS-regulated activities by hydrolyzing or altering N-acyl homoserine lactone (AHL) signaling molecules and auto-inducing peptides (AIPs) [46]. Quorum quenching enzymes such as lactonases usually carry out hydrolysis of AHLs which helps in preventing pathogenicity and biofilm development as shown in Figure 3 [47]. As members of the metallo-Β-lactamase superfamily, these enzymes usually depend on a dinuclear zinc site for stability and catalysis [48,49]. Research on lactonases such as GcL, AiiB, and AiiA has demonstrated their high catalytic efficiency and wide substrate specificity [47,48]. Key active site characteristics, such as metal coordination and substrate binding modes, have been found by structural investigations, offering insights into their catalytic process [47,49]. Despite having comparable reaction pathways, these enzymes differ in their catalytic efficiency depending on the substrate and the enzyme. This is mostly because of changes in their acyl-binding cavities and molecular gate dynamics [50]. Oxidoreductases use pyrroloquinoline quinone (PQQ), a redox cofactor. In respiratory activities, it is essential for succinate: quinone oxidoreductases, which relate succinate oxidation to quinone reduction [51]. Oxidoreductases alter the 3-oxo group of AHLs, acylases break the amide link, and lactonases hydrolyze the lactone ring [43]. Both QS and non-QS germs include QQ enzymes, which can reduce pathogenic traits without causing bacterial death and restricting cell development [52]. Numerous uses of QQ have been investigated, including as probiotics in aquaculture, biocontrol agents for crop protection, and biofouling control in membrane bioreactors [45].
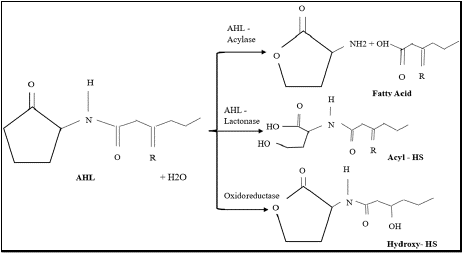
Figure 3: Effect of AHL- degrading enzymes on Acyl – Homoserine
lactones (AHLs).
Two – Component Signalling Systems
For bacteria to adapt to changes in their environment and stress reactions, two-component signaling systems (TCSs) are essential [53]. In most cases, these consist of a response regulator (RR) and a sensor histidine kinase (HK) [54]. Certain signals are picked up by HKs, which cause autophosphorylation and phosphor transfer to the corresponding RR, which frequently controls gene transcription [54,55]. The two-component system (TCS) is crucial for various biological processes, including the formation of the cell envelope, cellular proliferation and advancement of the cell cycle [55].
Sensor Histidine Kinases (HKs) in bacterial TCS systems, display a variety of domain configurations for both transduction and signal perception. These kinases usually have a conserved cytoplasmic kinase domain and a flexible input domain [56]. Periplasmic, transmembrane, or cytoplasmic input domains allow the cells to respond to a variety of external stimuli [57]. HKs are typically designed as modular proteins, and their sensor periplasmic domains are similar in structure to those periplasmic binding proteins [58].
Characteristically, Response Regulators (RRs) typically possess a variable C-terminal effector domain along with a conserved N terminal receiver domain [59]. The conformational changes obtained by phosphorylating the receiver domain at a conserved aspartic acid position activate the protein [60]. Although many RRs are transcription regulators, they employ a variety of regulatory techniques. When phosphorylated, certain RRs separate from DNA, while others form homodimers or heterodimers [59]. The effector domain may be capable of catalysis, protein binding, RNA binding, or DNA binding as shown in Figure 4 [59].
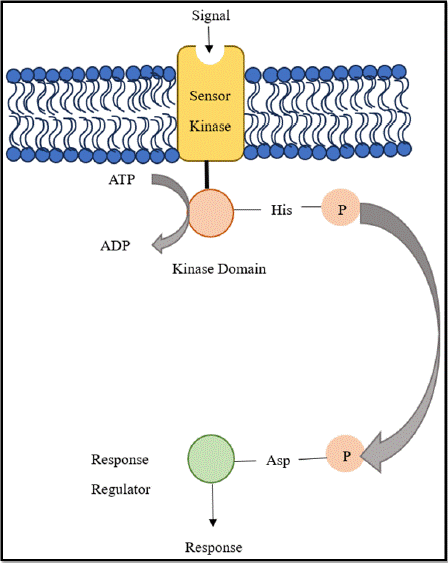
Figure 4: Diagrammatic representation of general Two-Component System
in Bacteria.
Mechanism: The SK uses its input domain, which can be periplasmic, transmembrane, or cytoplasmic, depending on the type of signal, to detect particular stimuli [56]. When activated, the SK undergoes a change in conformation that brings out a cellular response due to autophosphorylation at a histidine residue and the subsequent transfer of this phosphoryl group to an aspartate residue in the RR [61]. Depending on the particular HK, either cisor trans-autophosphorylation can cause this process [62]. Numerous biochemical pathways for signal detection and phosphorylation state regulation are displayed by SKs, which allow bacteria to detect and react to a broad variety of chemical and physical cues [63].
Since TCSs are found in bacteria but not in mammals, they could be targets for novel antibiotics [54,64]. It has been demonstrated that TCS inhibitors target the HK's catalytic domain, resulting in structural changes and aggregation [65].
Signaling Mechanisms in Fungi
To react to environmental stimuli and control development and pathogenicity, fungi use several common signaling pathways. Important pathways include cyclic AMP (cAMP) signalling, mitogen activated protein kinase (MAPK) signaling cascade along with the calcium calcineurin pathway [66,67,68]. Fungi can integrate various inputs and generate complex responses because these routes frequently interact and display functional pleiotropy [5].
cAMP/Protein Kinase A(PKA) Signalling Mechanism Pathway
This signaling pathway adjusts fungal metabolism to the host environment and is involved in nutrient sensing, specifically glucose [69]. cAMP signaling affects the generation of secondary metabolites, sporulation, growth, development, and stress tolerance in biocontrol fungi [70]. Adenylate cyclase, PKA and it’s subunits, heterotrimeric G proteins (which consists ofa, Β, and γ subunits), G protein coupled receptors (GPCRs), regulators of G protein signaling (RGSs), downstream transcription factors, phospho-di-esterases and R as GTPases make up the pathway [70,71].
GPCRs are critical in controlling pathogenicity, nutrition detection, and pheromone sensing in fungi [72]. Activation of the cAMP-dependent PKA pathway regulates growth and metabolic processes that turn on or off according to external availability of nutrients [73]. Respective heterotrimeric G proteins are linked with transmembrane receptors and translate extracellular signals to intracellular effects [74].
Heterotrimeric Gproteins are crucial in regulating mating, pathogenicity, also, germination in fungi [70]. These G-proteins have a, Β, and γ t translate the signals from the receptors located at the cell surface to cytoplasmic effectors. When a ligand-bound receptor catalyzes the GDP-GTP exchange on the a subunit, G-proteins are activated. As a result, a-GTP and Βγ separate, which can both activate downstream effectors [75]. G-proteins govern the specificity and temporal features of cellular responses. The complexity of G-protein signaling is enhanced by the fact that some species of fungi possess noncanonical GΒ subunits, which interact with unique Ga subunits [76].
The pathway's key component, AC, transforms ATP into cAMP [70]. MgATP binds to the bilobal structure formed by the catalytic domains of adenylate cyclase, which has a deep cleft as shown in Figure 5.
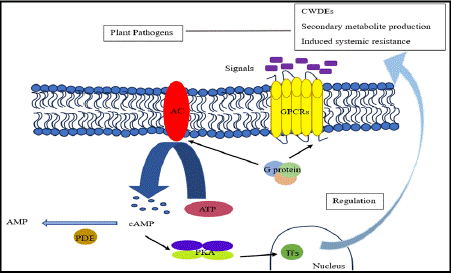
Figure 5: Mechanism of cAMP / PKA Signalling pathway in biocontrol fungi.
This pathway also involves multiple downstream transcription factors.
Mechanism: The route starts with GPCRs, which, when exposed to external stimuli like glucose, activate heterotrimeric G-proteins. These G-proteins next trigger adenylate cyclase (AC), which generates cAMP [70,77]. In fungi, cAMP activates Protein kinase A (PKA), which phosphorylates downstream transcription factors (TFs) that control gene expression [78]. G-proteins also affect cAMP levels in fungus, either promoting or suppressing AC, which is necessary for mating and pathogenicity. All fungal species share this pathway, which regulates vital processes like metabolism and the cell cycle while displaying species-specific adaptations in areas like the synthesis of secondary metabolites [79].
Mitogen Protein Kinase (MAPK) Signaling Pathway in Fungi
MAPK cascades regulate numerous functions, such as cell cycle regulation, cell division, mitogenesis, mating, morphogenesis, stress reactions, and virulence [80]. In response to host and environmental cues, plant pathogenic fungi use conserved MAPK pathways to control infection and development [81]. Fungal MAPKs facilitate host tissue penetration during plant-fungal interactions. Thus, it can be said, that MAPK cascades contribute to interrelated signaling events, resulting in a complex molecular dialogue [82].
Mechanism: MAPK signaling can be initiated either by G proteincoupled receptors (GPCRs) and / or receptor tyrosine kinases (RTKs) through linked pathways. Through a sequence of protein interactions including Shc, Grb2, Sos, and Ras, RTKs activate MAPK. GPCRs can also activate MAPK through GΒγ subunits, which can cause Shc to become tyrosine phosphorylated and form the Shc-Grb2-Sos complex as shown in Figure 6.
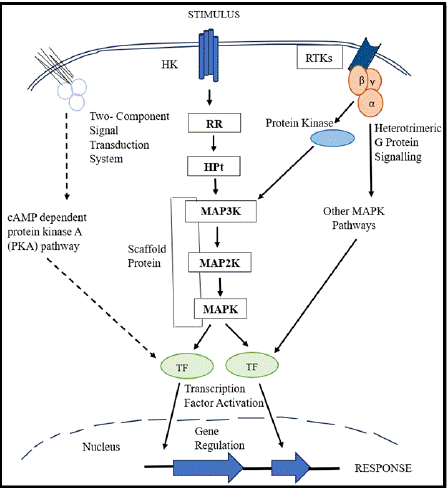
Figure 6: MAPK Signaling Cascade in Fungi.
Three tiers—MAP3K, MAP2K, and MAPK—make up the MAPK signaling cascade, where they interact successively through phosphorylation events thus, in turn, controlling cellular functions and sending a variety of extracellular signals [83]. Scaffold proteins assist the connections between these kinases by arranging the MAPK module into a multienzyme complex and adjusting the degree of signaling [84]. MAP3K and MAP2K catalyze consecutive dual phosphorylation events; MAP3K exhibits Ser/Thr kinase activity, whereas MAP2K has dual-specificity kinase activity [85]. Cells have delayed responses and steep signal-to-response curves as an effect of this organized mechanism [85].
Calcium-Calcineurin Signalling Pathway
Calcium-calcineurin signaling pathway plays a major role in a fungi-mediated survival, virulence and resistance toward therapies [86,87].
Mechanistic studies have localized two calcium influx systems, the low-affinity Ca2+ influx system (LACS) and the high-affinity Ca2+ influx system (HACS) [88]. HACS includes the Cch1, Mid1 and Ecm7 proteins, while LACS is independent of these components [88,89]. In fungi, this pathway consists of calcium channels, pumps, transporters, and their associated proteins [86]. The pathway consists of the calcium-calmodulin-dependent phosphatase, calcineurin and its downstream transcription factor, Crz1 [90,91].
In fungal cells, the highly conserved calcium-binding protein, calmodulin (CaM) regulates intracellular activity by triggering calcineurin and other enzymes [92].
Calcineurin (CN) is a unique highly conserved Ca2+- and calmodulin-dependent serine/threonine protein phosphatase that consists of a catalytic subunit (calcineurin A) and a regulatory subunit (calcineurin B) [93]. Calcineurin regulates multiple virulence determinants in human fungal infections including morphogenesis, tolerance to antifungals, growth at host temperature and sexual reproduction [94].
The transcription factor Crz1 regulates various cellular functions in fungi, including virulence, cell wall integrity, calcium homeostasis and stress response [91,94]. Calcineurin dephosphorylates Crz1 allowing for its transport to the nucleus and gene regulation [95].
The correct localization and function of Crz1 depend on it’s intrinsically disordered regions [95].
A crucial element of this pathway is Prz1, a zinc finger transcription factor that is translocated from the cytoplasm to the nucleus upon activation by calcineurin-mediated dephosphorylation [96]. Prz1 controls many genes, including the Ca2+ pump Pmc1, which is especially important in calcium homeostasis and cell wall production [96,97].
This pathway usually regulates several significant Ca2+-dependent target genes, including PMC1, PMR1, and PMR2. While PMC1 encodes a vacuolar Ca2+ ATPase, PMR1 and PMR2 encode P-type ion pumps linked to Mn2+ and Na+ tolerance, respectively. Through the transcription factor Crz1p, calcineurin positively controls the expression of these genes. Calcineurin also blocks the vacuolar H+/ Ca2+ exchanger VCX1, primarily via post-translational mechanisms as shown in Figure 7.
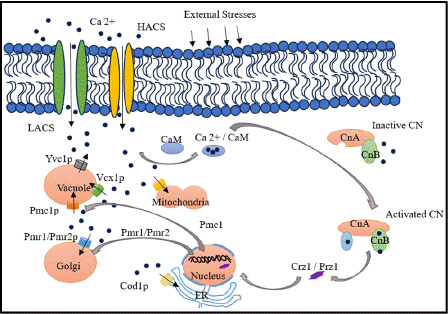
Figure 7: Calcium- Calcineurin Signaling Mechanism in Fungi.
This pathway usually regulates several significant Ca2+-dependent target genes, including PMC1, PMR1, and PMR2. While PMC1 encodes a vacuolar Ca2+ ATPase, PMR1 and PMR2 encode P-type ion pumps linked to Mn2+ and Na+ tolerance, respectively. Through the transcription factor Crz1p, calcineurin positively controls the expression of these genes. Calcineurin also blocks the vacuolar H+/ Ca2+ exchanger VCX1, primarily via post-translational mechanisms as shown in Figure 7.
This pathway usually regulates several significant Ca2+-dependent target genes, including PMC1, PMR1, and PMR2. While PMC1 encodes a vacuolar Ca2+ ATPase, PMR1 and PMR2 encode P-type ion pumps linked to Mn2+ and Na+ tolerance, respectively. Through the transcription factor Crz1p, calcineurin positively controls the expression of these genes. Calcineurin also blocks the vacuolar H+/ Ca2+ exchanger VCX1, primarily via post-translational mechanisms as shown in Figure 7.
Conclusion
Out of all the features we can comprehend regarding signaling mechanisms in microbes, one of the most important features is how these cellular processes intricately link these microbial communities to the environment. Not only that, signalling mechanisms in bacteria and fungi allow them to communicate with each other via chemical means which in turn permit the microbes to exist in the environment as communities. They aid in regulating several functions of microbes including pathogenicity, biofilm formation, their ability to sense their environment and adapt to stressful conditions.
We have realized that studying signaling mechanisms in microbes can provide valuable insights into their cellular processes. This knowledge can help us understand how these processes may lead to a variety of diseases in higher organisms, including plants, animals, and humans.
Even though humans are actively studying these pathways, scientists are trying to develop new medications, including antibiotics, to treat bacterial and fungal diseases. Several strategies are now exploited to combat diseases by antagonizing the signaling mechanisms of microbes against each other.
Bacterial signaling mechanisms have been found to have applications in biotechnology, particularly in wastewater treatment and biodegradation. Similarly, fungal signaling mechanisms have shown promising results in the development of fungal-specific drugs and biomolecules, which can be utilized in the pharmaceutical, food, and agricultural industries.
However, these innovations are still in development and are lately under investigation for their potential benefits to humanity's future.
Declarations
Following statements are articulated for the commitment regarding this article.
Acknowledgement
Commenced with profound gratitude and reverence, I would like to extend my heartfelt appreciation to all in my esteemed institution, Techno India University, West Bengal whose unwavering support and guidance have been instrumental in the fruition of this endeavour.
First and foremost, I am indeed indebted to my esteemed supervisor, Dr.Malavika Bhattacharya whose sagacious counsel and unparalleled expertise have paved the path of my scholarly journey. Her guidance, support and valuable insights throughout this research venture have been instrumental in shaping the direction of the work.
I would also like to offer my immense gratitude to my cosupervisor, Sudeshna Sengupta, a brilliant research scholar of my esteemed university whose insightful feedback, scholarly acumen, constant support and guidance have helped me shape the direction of my work.
A special note of thanks to the Chancellor of Techno India University for providing the Laboratory facility & the staff administrators here, whose support behind the scenes have provided a nurturing and conducive environment for the whole academic pursuit.
I conclude on a thanking note to all with unfeigned guidance, who have walked me through every challenge paving the path to usher in triumph in this scholarly voyage.
Author’s Contribution
A.A: Manuscript Writing, R.K: Editing and Formatting, D.S: Editing and Proof Reading, S.S: Finalizing, M.B: Guiding and Supervising.
Data Availability
All graphical data provided in this article are self-generated by the author. All data provided through the theoretical expansion are cited properly.
References
- Flint HJ. Micro-organisms and the Microbiome. Why Gut Microbes Matter. 2020; 1-8.
- Abdulkarem AT, Hasan AA, Khudhur HR & Abed SM. Overview of Opportunistic Bacteria. Karbala Journal of Pharmaceutical Sciences. 2024.
- Little CR, Carris LM & Stiles CM. Introduction to Fungi. 2012.
- Islam E, Khan MT & Irem S. Biochemical mechanisms of signaling: perspectives in plants under arsenic stress. Ecotoxicology and environmental safety. 2015; 114: 126-133.
- Jacob S, Bühring S & Bersching K. Recent Advances in Research on Molecular Mechanisms of Fungal Signaling. Encyclopedia. 2022.
- Zhang L & Dong Y. Quorum sensing and signal interference: diverse implications. Molecular Microbiology. 2005; 53.
- Das S, Das S & Ghangrekar MM. Bacterial signalling mechanism: An innovative microbial intervention with multifaceted applications in microbial electrochemical technologies: A review. Bioresource technology. 2021; 126218.
- Miller MB & Bassler BL. Quorum sensing in bacteria. Annual review of microbiology. 2001; 55: 165-199.
- Deep A, Chaudhary UB & Gupta V. Quorum sensing and Bacterial Pathogenicity: From Molecules to Disease. Journal of Laboratory Physicians. 2011; 3: 4-11.
- Romero M, Acuna LM & Otero A. Patents on quorum quenching: interfering with bacterial communication as a strategy to fight infections. Recent patents on biotechnology. 2012; 6: 2-12.
- Du Y, Li T, Wan Y, Long Q & Liao P. Signal molecule-dependent quorumsensing and quorum-quenching enzymes in bacteria. Critical reviews in eukaryotic gene expression. 2014; 22: 117-132.
- Dong Y & Zhang L. Quorum sensing and quorum-quenching enzymes. Journal of microbiology. 2005; 43: 101-109.
- Salazar ME & Laub MT. Temporal and evolutionary dynamics of twocomponent signaling pathways. Current opinion in microbiology. 2015; 24: 7-14.
- Kozubowski L, Lee SC & Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cellular Microbiology. 2009; 11.
- Williams P, Winzer K, Chan WC & Camara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philosophical Transactions of the Royal Society B: Biological Sciences. 2007; 362: 1119- 1134.
- Whiteley M, Diggle SP & Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017; 551: 313-320.
- Lixa C, Mujo A, Anobom CD & Pinheiro AS. A structural perspective on the mechanisms of quorum sensing activation in bacteria. Anais da Academia Brasileira de Ciencias. 2015; 87: 2189-2203.
- Li Z & Nair SK. Quorum sensing: How bacteria can coordinate activity and synchronize their response to external signals? Protein Science. 2012; 21.
- Ruzheinikov SN, Das S, Sedelnikova S, Hartley A, Foster SJ, Horsburgh M, et al. The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS. Journal of molecular biology. 2001; 313: 111-122.
- Hilgers MT & Ludwig M. Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proceedings of the National Academy of Sciences of the United States of America. 2001; 98: 11169 - 11174.
- Schauder S, Shokat KM, Surette MG & Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Molecular Microbiology. 2001; 41.
- Rajan R, Zhu JG, Hu X, Pei D & Bell CE. Crystal structure of S-ribosylhomocysteinase (LuxS) in complex with a catalytic 2-ketone intermediate. Biochemistry. 2005; 44: 3745-3753.
- Rao RM, Pasha SN & Sowdhamini R. Genome-wide survey and phylogeny of S-Ribosylhomocysteinase (LuxS) enzyme in bacterial genomes. BMC Genomics. 2016; 17.
- Sengstack J, Aquino AD, Schilling KH, Davies J, Lehrer J, Ng CM, et al. The Preparation and Screening of Several Novel Bacterial Homoserine Lactone Autoinducers. The FASEB Journal. 2016; 30.
- Beveridge T. Use of the Gram stain in microbiology. Biotechnic & Histochemistry. 2001; 76: 111-118.
- Desvaux M, Dumas É, Chafsey I & Hébraud M. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS microbiology letters. 2006; 256: 1-15.
- Banerjee G & Ray AK. Quorum-sensing network-associated gene regulation in Gram-positive bacteria. Actamicrobiologica et immunologica Hungarica. 2017; 64: 439-453.
- Monnet V & Gardan R. Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide’ Molecular Microbiology. 2015; 97.
- Novick RP &Geisinger E. Quorum sensing in staphylococci. Annual review of genetics. 2008; 42: 541-564.
- Le KY & Otto M. Quorum-sensing regulation in staphylococci—an overview. Frontiers in Microbiology. 2015; 6.
- Thoendel MJ, Kavanaugh JS, Flack CE & Horswill AR. Peptide signaling in the staphylococci. Chemical reviews. 2011; 111: 117-151.
- Taskar, K., & Gupta, S. A Review: Exploring Basic Methods of Gram Positive and Gram-Negative Bacteria. Invertis Journal of Renewable Energy. 2021; 11: 33-38.
- Pacheco, A.G., Alcântara, A.F., Abreu, V.G., & Corrêa, G.M. Relationships Between Chemical Structure and Activity of Triterpenes Against Gram- Positive and Gram-Negative Bacteria. 2012.
- Martin, C.A. Gram-Negative Bacteria. 2010.
- Papenfort, K., &Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nature Reviews Microbiology. 2016; 14: 576-588.
- Bosgelmez-Tinaz, G. Quorum Sensing in Gram-Negative Bacteria. Turkish Journal of Biology. 2003; 27: 85-93.
- Wu, H., Song, Z., Høiby, N., & Givskov, M. Quorum sensing in Gram-negative bacteria. Progress in Natural Science. 2004; 14: 377-387.
- Atkinson, S., Cámara, M., & Williams, P.R. N-Acylhomoserine Lactones, Quorum Sensing and Biofilm Development in Gram-negative Bacteria. 2016.
- Myszka, K., &Czaczyk, K. N-Acylhomoserine Lactones (AHLs) as Phenotype Control Factors Produced by Gram-Negative Bacteria in Natural Ecosystems. 2012.
- Smith, D.S., Wang, J., Swatton, J.E., Davenport, P.W., Price, B., Mikkelsen, H., et al. Variations on a Theme: Diverse N-Acyl Homoserine Lactone- Mediated Quorum Sensing Mechanisms in Gram-Negative Bacteria. Science Progress. 2006; 89: 167-211.
- Churchill, M.E., & Chen, L. Structural basis of acyl-homoserine lactonedependent signaling. Chemical reviews. 2011; 111: 68-85.
- Schuster, M., & Greenberg, E.P. LuxR-Type Proteins in Pseudomonas aeruginosa Quorum Sensing: Distinct Mechanisms with Global Implications. 2008.
- Wang, L., Dong, Y., & Zhang, L. Quorum Quenching: Impact and Mechanisms. 2008.
- Zhu, X., Chen, W., Bhatt, K., Zhou, Z., Huang, Y., Zhang, L., Chen, S., & Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Frontiers in Plant Science. 2023; 13.
- Fetzner, S. Quorum quenching enzymes. Journal of biotechnology. 2015; 201: 2-14.
- Czajkowski, R., &Jafra, S. Quenching of acyl-homoserine lactone-dependent quorum sensing by enzymatic disruption of signal molecules. Actabiochimica Polonica. 2009; 56: 1-16.
- Bergonzi, C., Schwab, M., Naik, T.V., & Elias, M.H. The Structural Determinants Accounting for the Broad Substrate Specificity of the Quorum Quenching Lactonase GcL. Chem Bio Chem. 2019; 20.
- Liu, D., Thomas, P.W., Momb, J., Hoang, Q.Q., Petsko, G.A., Ringe, D., & Fast, W. Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry. 2007; 46: 11789-11799.
- Liu, D., Momb, J., Thomas, P.W., Moulin, A.G., Petsko, G.A., Fast, W., & Ringe, D. Mechanism of the Quorum-Quenching Lactonase (AiiA) from Bacillus thuringiensis. 1. Product-Bound Structures. Biochemistry. 2008; 47: 7706-7714.
- Surpeta, B., Grulich, M., Palyzová, A., Marešová, H., & Brezovsky, J. Common Dynamic Determinants Govern Quorum Quenching Activity in N-terminal Serine Hydrolases. bioRxiv. 2022.
- Lancaster, C.R. Succinate: Quinone Oxidoreductases. 2011.
- Chen, F., Gao, Y., Chen, X., Yu, Z., & Li, X. Quorum Quenching Enzymes and Their Application in Degrading Signal Molecules to Block Quorum Sensing- Dependent Infection. International Journal of Molecular Sciences. 2013; 14: 17477-17500.
- Laub, M.T. The Role of Two-Component Signal Transduction Systems in Bacterial Stress Responses. 2011.
- Wang, S. Bacterial Two-Component Systems: Structures and Signaling Mechanisms. 2012.
- Skerker, J.M., Prasol, M.S., Perchuk, B.S., Biondi, E.G., & Laub, M.T. Two- Component Signal Transduction Pathways Regulating Growth and Cell Cycle Progression in a Bacterium: A System-Level Analysis. PLoS Biology. 2005; 3.
- Mascher, T., Helmann, J.D., & Unden, G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiology and Molecular Biology Reviews. 2006; 70: 910 – 938.
- Ishii, E., & Eguchi, Y. Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases. Biomolecules. 2021; 11.
- Cheung, J., Le-Khac, M., & Hendrickson, W.A. Crystal structure of a histidine kinase sensor domain with similarity to periplasmic binding proteins. Proteins: Structure. 2009; 77.
- Li, L., Wei, K., Jiang, W., & Lu, Y. Diversity of regulatory strategies for twocomponent system response regulators in bacteria. 2017.
- Bent, C.J., Isaacs, N.W., Mitchell, T.J., & Riboldi-Tunnicliffe, A. Crystal Structure of the Response Regulator 02 Receiver Domain, the Essential YycF Two-Component System of Streptococcus pneumoniae in both Complexed and Native States. Journal of Bacteriology. 2004; 186: 2872 - 2879.
- Stock, A.M., Robinson, V.L., & Goudreau, P.N. Two-component signal transduction. Annual review of biochemistry. 2000; 69: 183-215.
- Casino, P., Rubio, V., & Marina, A. The mechanism of signal transduction by two-component systems. Current opinion in structural biology. 2010; 20: 763-771.
- Krell, T., Lacal, J., Busch, A.E., Silva-Ji´menez, H., Guazzaroni, M., & Ramos, J.L. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annual review of microbiology. 2010; 64: 539-559.
- Bekker, M., Teixeira de Mattos, M.J., & Hellingwerf, K.J. The Role of Two- Component Regulation Systems in the physiology of the Bacterial Cell. Science Progress. 2006; 89: 213 - 242.
- Stephenson, K., Yamaguchi, Y.D., & Hoch, J.A. The Mechanism of Action of Inhibitors of Bacterial Two-component Signal Transduction Systems*. The Journal of Biological Chemistry. 2000; 275: 38900 - 38904.
- Park, G., Jones, C.A., & Borkovich, K.A. Signal Transduction Pathways. 2010.
- D’souza, C., &Heitman, J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS microbiology reviews. 2001; 25: 349-364.
- Lengeler, K.B., Davidson, R.C., D’souza, C., Harashima, T., Shen, W., Wang, P., et al. Signal Transduction Cascades Regulating Fungal Development and Virulence. Microbiology and Molecular Biology Reviews. 2000; 64: 746 - 785.
- Caza, M., & Kronstad, J.W. The cAMP/Protein Kinase A Pathway Regulates Virulence and Adaptation to Host Conditions in Cryptococcus neoformans. Frontiers in Cellular and Infection Microbiology. 2019; 9.
- Sun, Z., Yu, S., Wang, C., & Wang, L. cAMP Signalling Pathway in Biocontrol Fungi. Current Issues in Molecular Biology. 2022; 44: 2622 - 2634.
- Park, H., Kim, M., Yu, J., & Shin, K. Heterotrimeric G-Protein Signalers and RGSs in Aspergillusfumigatus. Pathogens. 2020; 9.
- Xue, C., Hsueh, Y., & Heitman, J. Magnificent seven: roles of G proteincoupled receptors in extracellular sensing in fungi. FEMS microbiology reviews. 2008; 32: 1010-1032.
- Ribeiro, L.F., Chelius, C., Boppidi, K.R., Naik, N.S., Hossain, S., Ramsey, J.J., et al. Comprehensive Analysis of Aspergillusnidulans PKA Phosphorylome Identifies a Novel Mode of CreA Regulation. mBio. 2019; 10.
- Nadarajah, K.K., Kumar, I.S., Sangapillai, V., & Omar, N.S. Recent advances in understanding the fungal G-protein-coupled receptors. Malaysian Journal of Microbiology. 2018; 14: 611-623.
- Oldham, W.M., & Hamm, H.E. Structural basis of function in heterotrimeric G proteins. Quarterly Reviews of Biophysics. 2006; 39: 117 - 166.
- Hoffman, C.S. Propping Up Our Knowledge of G Protein Signaling Pathways: Diverse Functions of Putative Noncanonical GΒ Subunits in Fungi. Science’s STKE. 2007: pe3 - pe3.
- Tamaki, H. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. Journal of bioscience and bioengineering. 2007; 104: 245-250.
- Chen, S., Lin, H., & Hsueh, Y. The cAMP-PKA pathway regulates prey sensing and trap morphogenesis in the nematode-trapping fungus Arthrobotrysoligospora. G3: Genes| Genomes| Genetics. 2022; 12.
- Guo, L., Breakspear, A., Zhao, G., Gao, L., Kistler, H.C., Xu, J., & Ma, L. Conservation and divergence of the cyclic adenosine monophosphate– protein kinase A (cAMP–PKA) pathway in two plant-pathogenic fungi: Fusariumgraminearum and F. verticillioides. Molecular Plant Pathology. 2015; 17: 196 – 209.
- Martínez-Soto, D., & Ruiz-Herrera, J. Functional analysis of the MAPK pathways in fungi. Revistaiberoamericana de micologia. 2017; 34: 192-202.
- Jiang, C., Zhang, X., Liu, H., & Xu, J. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathogens. 2018; 14.
- Hamel, L., Nicole, M., Duplessis, S., & Ellis, B.E. Mitogen-Activated Protein Kinase Signaling in Plant-Interacting Fungi: Distinct Messages from Conserved Messengers[W]. Plant Cell. 2012; 24: 1327 – 1351.
- Keshet, Y., & Seger, R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods in molecular biology. 2010; 661: 3-38.
- Dhanasekaran, D.N., Kashef, K., Lee, C.M., Xu, H., & Reddy, E.P. Scaffold proteins of MAP-kinase modules. Oncogene. 2007; 26: 3185-3202.
- Piala, A.T., Humphreys, J.M., & Goldsmith, E.J. MAP kinase modules: the excursion model and the steps that count. Biophysical journal. 2014; 107: 2006-2015.
- Liu, S., Hou, Y., Liu, W., Lu, C., Wang, W., & Sun, S. Components of the Calcium-CalcineurinSignaling Pathway in Fungal Cells and Their Potential as Antifungal Targets. Eukaryotic Cell. 2015; 14: 324 - 334.
- Li, W., Shrivastava, M., Lu, H., & Jiang, Y. Calcium-calcineurinsignaling pathway in Candida albicans: A potential drug target. Microbiological research. 2021; 249: 126786.
- Muller, E.M., Locke, E.G., & Cunningham, K.W. Differential regulation of two Ca(2+) influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics. 2001; 159: 1527-1538.
- Martin, D., Kim, H., Mackin, N.A., Maldonado-Báez, L., Evangelista, C., Beaudry, V., et al. New Regulators of a High Affinity Ca2+ Influx System Revealed through a Genome-wide Screen in Yeast*. The Journal of Biological Chemistry. 2011; 286: 10744 - 10754.
- Park, H., Lee, S.C., Cárdenas, M.E., & Heitman, J. Calcium-Calmodulin- CalcineurinSignaling: A Globally Conserved Virulence Cascade in Eukaryotic Microbial Pathogens. Cell host & microbe. 2019; 26: 453-462.
- Yang, Y., Xie, P., Li, Y., Bi, Y., & Prusky, D. Updating Insights into the Regulatory Mechanisms of Calcineurin-Activated Transcription Factor Crz1 in Pathogenic Fungi. Journal of Fungi. 2022; 8.
- Juwadi, P.R., & Chivukula, S. The emerging role of Calcium and Calmodulin in the regulation of fungal growth, development and metabolism. 2004.
- Rusnak, F.M., & Mertz, P.S. Calcineurin: form and function. Physiological reviews. 2000; 80: 1483-1521.
- Zhang, T., Xu, Q., Sun, X., & Li, H. The calcineurin-responsive transcription factor Crz1 is required for conidation, full virulence and DMI resistance in Penicilliumdigitatum. Microbiological research. 2013; 168: 211-222.
- Chadwick, B.J., Ross, B.E., & Lin, X. Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans. Journal of Fungi. 2023; 9.
- Hirayama, S., Sugiura, R., Lu, Y., Maeda, T., Kawagishi, K., Yokoyama, M., et al. Zinc Finger Protein Prz1 Regulates Ca2+ but Not Cl- Homeostasis in Fission Yeast. The Journal of Biological Chemistry. 2003; 278: 18078 – 18084.
- Chatfield-Reed, K., Vachon, L., Kwon, E.G., & Chua, G. Conserved and Diverged Functions of the Calcineurin-Activated Prz1 Transcription Factor in Fission Yeast. Genetics. 2016; 202: 1365 – 1375.
- Bonilla, M., Nastase, K., & Cunningham, K.W. Essential role of calcineurin in response to endoplasmic reticulum stress. The EMBO Journal. 2002; 21.
- Yoshimoto, H., Saltsman, K.A., Gasch, A.P., Li, H.X., Ogawa, N., Botstein, D., et al. Genome-wide Analysis of Gene Expression Regulated by the Calcineurin/Crz1p Signaling Pathway in Saccharomyces cerevisiae *. The Journal of Biological Chemistry. 2002; 277: 31079 - 31088.
- Maraz, K.M., & Khan, R.A. An overview on impact and application of microorganisms on human health, medicine and environment. GSC Biological and Pharmaceutical Sciences. 2021.