
Editorial
J Bacteriol Mycol. 2014;1(1): 2.
Delineating the Oxidative Stress Response in Shewanella Oneidensis
Jie Yuan and Haichun Gao
Institute of Microbiology, Zhejiang University, China
*Corresponding author: Haichun Gao, Institute of Microbiology and College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang, 310058, China.
Received: Aug 16, 2014; Accepted: Aug 18, 2014; Published: Aug 18, 2014
Editorial
Emergence of the ability to respire on O2 in prokaryotes had been a great evolutionary success story, leading way to more efficient energy use, faster growth, greater cell populations for natural selection to act upon, and eventually to the birth of higher organisms [1]. However, every coin has two sides, along with all the benefits of aerobic respiration come the Reactive Oxygen Species (ROS) and oxidative stress [2].
Superoxide (O2 -), H2O2 and Organic Peroxides (OP) are all ROS species commonly encountered by microbes [3,4]. They are damaging to DNA, RNA, protein, lipids, and virtually any cellular component, causing oxidative stress [2, 5]. The bad news is, beside radiation and other external factors, one major source of ROS is the process of aerobic respiration per se, rendering ROS an unavoidable drag of the aerobic lifestyle [1,5]. In defense, microorganisms have evolved sophisticated mechanisms to sense, respond, and battle against ROS.
Figure 1 summarizes the oxidative stress response dedicated to battling against various ROS and their damages. Although a diversity of genes are involved, and specific genes differ for different ROS species, oxidative stress response can be broadly divided into two tiers of defense. The primary defense focuses on removing the ROS (e.g., H2O2 induces catalase and peroxidase, and O2- induces superoxide dismutase), and the secondary defense is concerned with cellular component repair or removal [4,5]. Also illustrated in Figure 1 are the four major sensor-transcriptional regulators well characterized in model organisms, OxyR, PerR, OhrR, and SoxRS. As shown, each of these regulators specializes in coping with a set of different ROS species, with OxyR and PerR for H2O2 but usually found in different organisms, OhrR for OPs, and SoxRS for superoxide.
While studies in model organisms undoubtedly helped us put together a general picture of oxidative stress response, it is also of great value to expand the territory by characterizing other interesting organisms, especially those thriving in redox-stratified environments prone to ROS generation, and investigate their specific strategies. Shewanella oneidensis MR-1, a gram-negative facultative anaerobe, is such a representative. Endowed by the diverse collection of iron or heme containing respiratory proteins encoded in its genome, MR-1 is able to respire on oxygen, nitrate, Fe(III), Cr(VI) and even more exotic electron acceptors [6]. These characteristics all put MR-1 in the front seat for ROS attack [2,5], and our study aims to shed light on how this organism defends itself. We have identified analogues of OxyR and OhrR MR-1 [7,8]. And our recent research had revealed that the regulation of oxidative stress response in MR-1 (summarized in Figure 2) differ considerably from the general model shown in Figure 1.
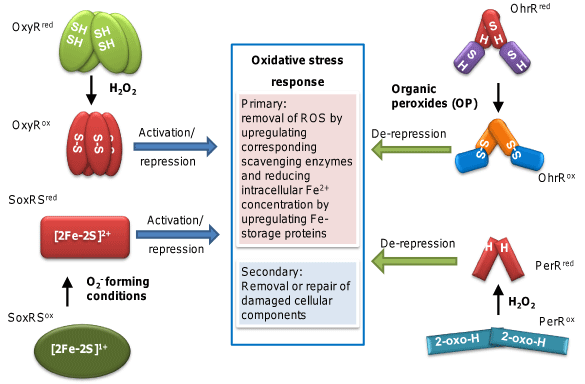
Figure 1: Summary of general oxidative stress response and known major
ROS sensor-transcriptional regulators. OxyR is the principle H2O2-responding
regulator governing the expression of a similar set of genes (OxyR regulon)
across species. OxyR features a key Cysteine residue (C199 in E. coli) prone
to H2O2 oxidation, although still a point of debate, this oxidation most likely
results in the formation of a disulfide bond (between C199 and C208 in E.
coli) which leads to OxyR activation and eventually increased expression of
genes for scavenging enzymes such as peroxidase and catalase (Ahp and
KatG) and other proteins having a role in oxidative stress response. Although
OxyR is widely distributed, Bacillus subtilis employs PerR in its place. PerR is
a repressor that can be deactivated by H2O2 through oxidation of a histidine
residue, its regulon overlaps to a great extent with OxyR. It is interesting to
contemplate how such distinct H2O2-sensing mechanisms had evolved, but
no compelling hypothesis had been put forward. SoxR senses superoxidegenerating
conditions (rather than O2- per se) through redox reaction of its
2Fe-2S clusters and often modulates oxidative stress response by regulating
the expression of its cognate transcriptional regulator SoxS. OhrR senses
OP in a manner similar to OxyR with H2O2, except that it functions mainly
as a repressor. In Xanthomonas campestris OhrR, OP-triggered formation
of the intermolecular disulfide between the sensory residue C22 and the
C127 residue of the other subunit of the homodimeric protein leads to the derepression
of Ohr. A subset of OhrR proteins known as Cys-1 contains only
one conserved sensing cysteine (e.g., C15 in B. subtilis). It forms either mixed
disulfide with cellular free thiol or cyclic sulfonamide with the amino group of
a nearby amino acid upon organic peroxide oxidation. For thorough reviews,
please refer to [3-5].
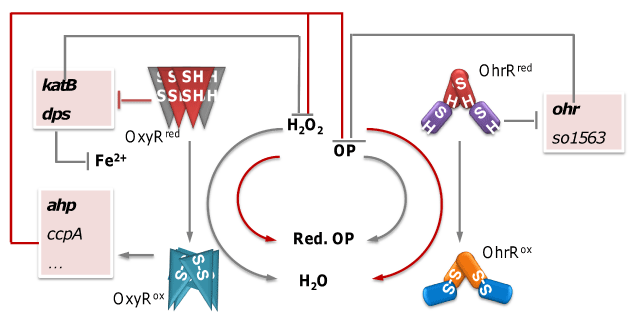
Figure 2: Summary of the oxidative stress response characterized in S. oneidensis MR-1 recently. Grey line indicates reactions or relationships existing in E. coli, and red lines indicate those that are in MR-1 but not E. coli.
S. oneidensis OxyR represents a relatively rare H2O2-responding regulator, which functions as both an activator and a repressor as in Neisseria [9]. Although three catalases (KatB, KatG-1, and KatG-2) are encoded in the genome, only KatB appears to be functional. In addition, several peroxidase genes of the OxyR regulon are under positive control of the regulator but their contribution in combating H2O2 is negligible. Instead, the predominant force protecting the microorganism from harmful H2O2 relies on de repression of the katB and dps genes upon inactivation of OxyR, whose products ensure rapid removal of the oxidant and restriction of intracellular iron concentrations, respectively [7]. One striking difference between the S. oneidensis OxyR regulon and those characterized to date is that OxyRSo seems not to be involved in the regulation of genes encoding proteins participating in the secondary defense, such as those for heme synthesis, FeS cluster assembly, or divalent cat ion import. This observation may partially account for the hypersensitivity of S. oneidensis (in comparison to E. coli) to H2O2 [7].
Loss of OxyRSo results in substantial defects in both growth and viability. While we do not yet know the mechanism underlying the slow growth, impaired viability is certainly associated with iron [7]. Addition of Fe3+ fully rescues the defect, whereas Mn2+ does not have such effect, despite that replacement of vulnerable Fe2+ by Mn2+ in many proteins is believed to be a critical strategy against oxidative stress in E. coli [5]. This observation is clearly out of expectation because extra iron is usually blamed for exasperating the stress via Fenton reaction, leading to generation of the most deadly ROS, hydroxyl radical. Moreover, OxyRSo cannot functionally complement OxyREc or vice versa. As far as we are aware, this is the first case that an OxyR analogue could not work as a functional replacement in E. coli.
The most distinctive feature we have observed in S. oneidensis is the cross talk between OxyR and OhrR systems [8]. More specifically, these two regulators can both respond to H2O2 as well as OPs; and Ahp contributes considerably to OP scavenging. Finally, it is also worth mentioning that the OhrR regulon of S. oneidensis differs from its characterized counterparts in that it at least contains one extra member SO1563 (glutathione peroxidase), in addition to Ohr (Organic hydroperoxide resistant protein), although its role in ROS stress response remains elusive.
These intriguingly unusual arrangement (in comparison to E. coli) and the observation that MR-1 is far more sensitive to H2O2 (but not to OP) than E. coli, prompted us to the postulation that organic peroxides are probably more of a threat than H2O2 in the natural habitat of MR-1. Given that E. coli usually dwells in human gut, it probably often encounters H2O2 attack from the host immune system [10]. In contrast, MR-1 most typically resides in aqueous environments relatively rich in organic matters and mineral. A recent report [11] had demonstrated that simple organic matters can be effectively converted to acyl hydro peroxides in the presence of H2O2 and minerals such as Fe2O3, Al2O3, TiO2 mineral, supporting our theory.
Last but not least, we would like to acknowledge that these findings just opened up the investigation of S. oneidensis oxidative stress response, the molecular detail of how OxyR and OhrR senses both types of ROS remains to be elucidated, whether the KatG-1 and KatG-2 are just useless evolutionary leftovers or a secret arsenal against some other ROS threat would be interesting, not to mention that despite owning a SOD in the genome, no Sox analogues was detected in MR-1. All these puzzles invite further studies to put together a complete roadmap of S. oneidensis oxidative stress response.
References
- 1. Madigan MT, et al. Toxic Forms of Oxygen, in Brock Biology of Microorganisms (13th Edition), D.E.e. al., Editor. 2012. Pearson Education, Inc. San Francisco, CA.
- 2. Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999; 2: 188-194.
- 3. Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol. 2012; 194: 5495-5503.
- 4. Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008; 77: 755-776.
- 5. Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013; 11: 443-454.
- 6. Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008; 6: 592-603.
- 7. Jiang Y, Dong Y, Luo Q, Li N, Wu G, Gao H. Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis. J Bacteriol. 2014; 196: 445-458.
- 8. Li N, Luo Q, Jiang Y, Wu G, Gao H. Managing oxidative stresses in Shewanella oneidensis: intertwined roles of the OxyR and OhrR regulons. Environ Microbiol. 2014; 16: 1821-1834.
- 9. Ieva R, Roncarati D, Metruccio MM, Seib KL, Scarlato V, Delany I. OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol Microbiol. 2008; 70: 1152-1165.
- 10. Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol. 2008; 15: 164-187.
- 11. Zhao Y, Huang D, Huang L, Chen Z. Hydrogen peroxide enhances the oxidation of oxygenated volatile organic compounds on mineral dust particles: a case study of methacrolein. Environ Sci Technol. 2014.