
Special Article - Breeding Techniques
Ann Agric Crop Sci. 2021; 6(4): 1084.
Impact of Stripe Rust Resistance Alleles on Wheat Grain Yield Using Landraces and Improved Accessions
ElBasyoni IS1,2*
¹Department of Crop Science, Faculty of Agriculture,Damanhur University, Damanhur, Egypt
²Department of Agronomy and Horticulture, University of Nebraska-Lincoln, Lincoln, NE, USA
*Corresponding author: Ibrahim S. ElBasyoni, Crop Science Department, Faculty of Agriculture, Damanhur University, Damanhur, Egypt
Received: May 11, 2021; Accepted: June 10, 2021; Published: June 17, 2021
Abstract
Stripe rust is one of the most devastating biotic stresses to cause grain yield losses in wheat. In the current study, 227 imported accessions, and six widely grown modern cultivars (Sids14, Sids12, Misr1, Misr2, Giza171, and Gimmiza9), were used. All plant materials were planted in the field and evaluated for stripe rust resistance and grain yield. Five Simple Sequence Repeats (SSR) markers Xpsp3000, Xbarc8, Xgwm419, Xwmc44, and Xbarc32, respectively, are associated with five essential stripe rust resistance genes Yr10, Yr15, Yr26, Yr29, and Yr59, were also used. The results indicated a highly positive and significant correlation between grain yield and stripe rust resistance. Furthermore, as the number of stripe rust resistance alleles increased, both grain yield and stripe rust resistance increased. Out of the 233 accessions used, 11 accessions were found to contain the five resistance genes. The identified resistant accessions could be used as a gene source to enhance stripe rust resistance in wheat breeding programs. SSR markers used in the current study effectively capture a substantial part of the phenotypic variation caused by stripe rust. Thus, these five markers could be used effectively in marker-assisted selection for stripe rust resistance.
Keywords: SSR; Yellow Rust; Wild Wheat Types; Genetic Diversity
Introduction
Wheat (Triticum aestivum L), the leading staple food crop worldwide, is often attacked by several fungal, bacterial, viral, and nematode pathogens. Wheat rusts (Puccinia spp.) are among the most devastating pathogens that attack wheat, causing significant yield losses [1]. The most common types of wheat rust are leaf rust caused by Puccinia triticina f.sp. tritici, stem rust caused by Puccinia graminis f.sp. tritici, and stripe rust caused by Puccinia striiformis f.sp. tritici [2]. Stripe rust is regarded as a deleterious wheat rust disease [3]. Therefore, stripe rust is considered the most critical hazard for global wheat production [4]. Stripe rust used to be a cool weatheradapted disease, but recently aggressive races have also spread to warm-weather parts of the world [5].
Several stripe rust pathogens originated from Europe, Australia, and North America, and some pathogen populations within these regions experience a high genetic diversity level [6]. Moreover, western China, Central Asia, and the Himalayas were considered the center of stripe rust pathogen evolution, where sexual recombination is common [7]. New stripe rust races that originated from the Himalayas region spread across Europe between 2011 and 2015 [6]. Egypt is one of the warm weather countries that recently suffered from stripe rust at high severity levels [8]. Grain yield losses between 14% and 26% due to stripe rust in the Nile Delta were recorded [8]. Shahin et al. [9] evaluate eight commercially grown Egyptian wheat cultivars for stripe rust under the field growth conditions. They concluded that most commercially grown wheat cultivars in Egypt possess low adult plant resistance levels to stripe rust.
Field evaluation for stripe rust has been a successful and effective way to identify stripe rust-resistant genotypes. However, for the field evaluation to be effective, a large number of accessions have to be grown and infected with several races or the predominant mixture of native races to identify resistant accessions. It also requires reliable rust screening nurseries [8]. To ensure the pathogenic races’ presence, it is desirable to inoculate the host plants with those races artificially [10]. But it is not acceptable to inoculate the host plants in the field with pathogenic races that are not naturally present in the evaluation environment [10]. Therefore, field evaluation for stripe rust is challenging because of the annual fluctuation of the climate conditions, which might also stimulate stripe rust evaluation and the presence of new races. Additionally, the traditional field evaluation methods are time-consuming. Therefore, several plant breeders have incorporated molecular markers in their breeding programs to identify and introgress rust resistance genes with minimal dragging effect into their elite lines [11].
In total, 81 stripe rust resistant genes were identified on various wheat chromosomes [1], designated Yr1 to Yr67, Yr73, and Yr74 [12,13]. Out of the 81 stripe rust-resistant genes, 18 are adult-plant resistance genes; Yr11-Yr14, Yr16, Yr18, Yr29, Yr30, Yr34, Yr36, Yr39, Yr46, Yr48, Yr49, Yr52, Yr54, Yr59, and Yr62, whereas 54 are seedling resistance genes [14]. Pyramiding adult and seedling resistant genes in a genotype could confer enhanced durable resistance to stripe rust of wheat [14]. The first step to pyramid several effective resistance genes is to identify genuinely resistant genotypes. Resistance genes can be tagged and identified rapidly and accurately using molecular markers in Marker-Assisted Selection (MAS). MAS have been used successfully to facilitate gene identification for crucial traits such as stripe, stem, and leaf rust resistance. Several DNA molecular marker platforms were commonly used successfully in MAS, such as RFLP [15], RAPD [16], SSR [17], SNP [18], and KASP [19]. SSR is the common molecular marker platform in MAS studies due to its reproducibility, multi-allelic nature, co-dominant inheritance, and robust amplification [20]. Simple Sequence Repeats (SSR) flanking genes that control a trait can be used to screen large populations for that trait in a short time. In addition, markers with diagnostic alleles, i.e., the size difference in the parental accessions, are ideal markers for MAS because they will be completely correlated with the trait [21]. Furthermore, MAS can be used in pyramiding adult and seedling resistant genes in a genotype [14].
Therefore, in the current study, we characterized a panel of 233 spring wheat accessions for stripe rust using five diagnostic SSR markers. The panel was also evaluated under the field conditions for two growing seasons in which stripe rust resistance and grain yield were measured. The current study’s objectives were to (1) screen the wheat collection for resistance to stripe rust and (2) estimate the impact of stripe rust favorable alleles accumulation on stripe rust resistance and grain yield production.
Materials and Methods
Germplasm
A panel comprised of 227 imported accessions and five widely grown modern cultivars (Sids14, Misr1, Misr2, Giza171, and Gimmiza9) were used in the current study. The imported wheat accessions contain 96 elite breeding lines, 74 cultivars, and 57 landraces. Thereafter, we will refer to the elite lines and cultivars as the improved accessions. The seeds of the imported accessions were collected by the USDA-ARS from several geographic regions around the world. In comparison, the commercial cultivars were obtained from the Agricultural Research Center (ARC), Egypt. For further information about the accessions details such as pedigrees and origin regions, the reader is referred to the supporting information Table S1.
Phenotyping
The spring wheat panel was phenotyped at Elbasotan region in an experimental farm for Damanhour University (30°46'46'' N, 30°82'32'' E) during two consecutive growing seasons 2018 and 2019. Grain yield (ton/hectare) and stripe rust score were measured on plots that were four 2.5-meter rows, 30cm apart. Plots were laid out in a randomized block design with three replicates per year. Planting dates were November 21st and November 14th for the first and second growing seasons, respectively. Stripe rust (incited by Puccinia striiformis f.sp) susceptible cultivar “Morocco” was planted around the experiment as a one-meter wide border. During the booting growth stage, Morocco was dusted with 200mg urediniospores of five prevalent and aggressive pathotypes of stripe rust, i.e., 0E0, 6E4, 70E20, 128E28, and 134E244, mixed with talcum powder (1:20, spores: talcum). Stripe rust was scored according to modified Cobb’s scale [22]. The infection type was expressed in the following classes, i.e., Immune = I, R = Resistant, small uredinia surrounded by necrosis; MR = Moderately Resistant, medium to large uredinia surrounded by necrosis; MS = Moderately Susceptible, medium to large uredinia surrounded by chlorosis; S = Susceptible, large uredinia without necrosis or chlorosis [23]. The statistical analysis was conducted on the infection types after converting it into 0, 2, 4, 6, and 8 for immune, resistant, moderately resistant, moderately susceptible, and susceptible, respectively. After physiological maturity, all plants in each plot were manually cut at 5cm above soil service and left to dry in the middle of the plots. Three days later, plants from each plot were threshed separately using a locally made single plot thresher, in which seeds were collected, weighed, and converted to tons/ha. Standard agronomic practices, including weed control, recommended nitrogen, phosphorus, and potassium applications, were followed.
Genotyping
The total genomic DNA was extracted from 200 mg of fresh leaves during the seedling stage. DNA extraction kit (Promega, USA) was used, and the manufacturer’s instructions were followed. The DNA concentration of each sample was measured using a spectrophotometer at a wavelength of 260 and 280 nm using a CARY 50 probe UV-visible spectrophotometer (Varian, CA, USA). The DNA quality was confirmed by running 5μl diluted DNA on a 0.8% agarose gel. Table 1 shows the specific band (bp) for stripe rust resistance for the SSR markers used in the current study. The primer sequences, linkage map location, and the amplification requirements for the SSR markers were obtained from the GrainGenes website (https://wheat.pw.usda.gov/cgi-bin/GG3/browse.cgi?class=marker). PCR products were scored as present (1) or absent (0) across the 233 accessions for each primer.
Marker
Gene
Product (bp)
Reference
Xpsp3000
Yr10
260
[54]
Xbarc8
Yr15
164
[55]
Xgwm419
Yr26
141
[56]
Xwmc44
Yr29
270
[57]
Xbarc32
Yr59
165
[49]
Table 1: SSR markers, along with their product size used to postulate stripe rust resistance genes in the studied wheat panel.
Statistical analysis
The following mixed-effect model [24] was used to estimate the Best Linear Unbiased Predictors (BLUP) for grain yield and stripe rust scores:
Yijm = μ + Ei + REj(i) + Gm + GEmi + ε ijm
where Yijm is the response, Ei is the ith level of years effect (Fixed), REj(i) is the effect of jth replicate nested within ith year (Fixed), Gm is the effect of mth accession (random), GEmi the interaction effect between mth accession and ith year (random) and εijlm is the random experimental error effect associated with Yijm.
Furthermore, the following parameters were estimated using the metan R package [24]. The genotypic effect (gi) for each accession was estimated as follows:
gi = h² g (yi.-y..)
where h² g is the broad-sense heritability, yi. and y.. are accession ith mean and the overall mean, respectively.
The predicted performance was estimated as follows: gi + μ
The Upper (UL) and Lower Limits (LL) of the predicted mean were estimated as follows:
gi + μ ± CI
where CI is the confidence interval, which was estimated as follows:
where t is the student’s t value for a two-tailed t test at a given probability error; AC is the accuracy of selection and s²g is the genotypic variance.
The broad-sense heritability (H²) was estimated as follows:
where s²G is the genetic variance, s²GxE is the genotype by year’s variance, and s²error is the residual variance.
Polymorphic information content (PIC) was calculated as follows:
where Pij and Pik are the frequencies of jth and kth alleles for marker i, respectively.
Cluster analysis was performed using the ape R package [26]. Since the studied variables, i.e., SSR markers, grain yield, and stripe rust scores, had different measurement units, all variables were standardized [27]. Then dendrogram was constructed utilizing Ward’s method based on squared Euclidean distance [28].
Means within and across years were compared using Tukey’s Honest Significant Difference (HSD) [29]. Homogeneity of variance across locations and years was tested using Bartlett’s Test [30]. The combined analyses of variance were then performed among traits with homogeneous variance across years, as outlined by Cochran and Cox [31].
Results
Analysis of variance and accessions performance
Bartlett’s test results indicated a homogeneous variance across years, in which the p-value was >0.05. Therefore, a combined analysis of variance across years was visible. The combined analysis of variance across years for grain yield and stripe rust is provided in Table 2. Results in Table 2 indicated a highly significant effect for accessions on grain yield and stripe rust resistance score. Furthermore, the interaction between accessions and years was not significant for grain yield. In comparison, the interaction between accessions and years was significant for stripe rust. The overall grain yield averaged across years and accessions was 5.6 tons/hectare and ranged from 3.9 to 7.5 tons/hectare.
Source
Df
MS
Yield
Stripe rust
Years
1
2.3
1.64
Replicates (Years)
4
4.42
90.3
Accessions
232
31.56**
31.1**
Accessions × Years
232
2.42ns
1.97*
Residuals
928
2.1
1.38
*, **: Significant at 0.05 and 0.01 probability levels, respectively. ns: non-significant.
Table 2: Analysis of variance for grain yield (tons/hectare) and stripe rust scores combined across years.
Interestingly, accession “PI 14392” ranked number 1, based on the accessions ranking across years with the average grain yield for PI 14392 at 7.47 tons/hectare. This was followed by Sids14, “PI 12782,” and “PI 312115,” in which they produced 7.31, 7.17, 7.15 tons/hectare, respectively (Table 3). In comparison, the lowest yielding accessions were “PI 304915”, “PI 192376”, and “PI 192294”, in which they produced 3.91, 3.96 and 3.99 tons/hectare, respectively. The Egyptian cultivars, Sids14, Sids12, Misr1, Misr2, Giza171, and Gimmiza9, had an average grain yield of 6.24 tons/hectare. Misr2 was the lowest yielding cultivars among the Egyptian ones, followed by Misr1, in which they produced 4.5 and 5.7 tons/hectare, respectively.
Grain yield tons/hectare
Accessions
Rank
Mean
BLUPg
Predicted
LL
UL
14392
1
7.6
1.9
7.5
7
7.9
Sids14
2
7.5
1.7
7.3
6.9
7.7
12782
3
7.3
1.6
7.2
6.7
7.6
312115
4
7.3
1.6
7.2
6.7
7.6
134136
5
7.3
1.6
7.1
6.7
7.6
1517
6
7.2
1.5
7.1
6.7
7.5
11254
7
7.2
1.5
7.1
6.7
7.5
190450
8
7.2
1.5
7.1
6.7
7.5
14244
9
7.2
1.5
7.1
6.7
7.5
193927
10
7.2
1.5
7.1
6.6
7.5
Stripe rust score
Accessions
Rank
Mean
BLUPg
Predicted
LL
UL
12782
1
0.7
-3
0.9
0.2
1.6
12811
2
0.7
-3
0.9
0.2
1.6
14282
3
0.7
-3
0.9
0.2
1.6
14362
4
0.7
-3
0.9
0.2
1.6
1517
5
0.7
-3
0.9
0.2
1.6
Giza171
6
0.7
-3
0.9
0.2
1.6
14249
7
1
-2.7
1.2
0.5
1.9
15035
8
1
-2.7
1.2
0.5
1.9
Sids14
9
1
-2.7
1.2
0.5
1.9
12782
10
0.7
-3
0.9
0.2
1.6
BLUPg is the genotypic effect. LL and UL are the upper (UL) and lower limits (LL) of the predicted mean.
Table 3: List and rank of the top 10 resistant accessions for stripe rust and ten of the highest yielding accessions.
Marker/Traits
Xbarc8
Xwmc44
Xbarc32
Xgwm419
Grain yield (% variance)
Stripe rust (% variance)
Xpsp3000
0.02ns
0.10ns
-0.10ns
0.34**
0.41** (16.9)
-0.55** (30.3)
Xbarc8
1
0.00ns
-0.07ns
0.17
0.35 *(12.6)
-4.824
Xwmc44
1
-0.32*
0.12
0.29* (8.3)
-2.436
Xbarc32
1
0.03
0.22* (4.8)
-0.558
Xgwm419
1
0.44** (19.8)
-0.47** (22.0)
Grain yield
1
-0.87
Stripe rust
1
Numbers in brackets refer to the % of variance explained by the marker for each trait. *, **: Significant at 0.05 and 0.01 probability levels, respectively. ns: non significant.
Table 4: Correlation among SSR markers, grain yield, and stripe rust.
Under the field conditions, a diverse adult plant reaction to the infection was observed, which was pronounced as a significant year by accession interaction. That significant year by accessions indicated that the accessions’ response to stripe rust varied from year to year. Based on the stripe rust score across years, the average score was 3.9 (on a stripe rust scale 0 to 8). The most resistant accessions were “PI 12782”, “PI 1517”, “PI 12811”, which all had an overall average score of 1 (Table 3). Conversely, the most susceptible accessions were “PI 220440”, “PI 192647”, and PI 225160”, in which they scored from 7 to 8 on the stripe rust resistance scale. Additionally, the Egyptian cultivars had an average score of 3.3, while Sids14 had an average overall score of 1.2, followed by Giza171 and Misr2, in which they had average scores of 0.9 and 4, respectively. The most susceptible Egyptian cultivars in the current study were Misr1 and Gimmiza9, in which they scored 5.9 and 4.6, respectively. The broad-sense heritability for grain yield was 62% and 57% for stripe rust scores.
Accessions characterization
In the current study, the SSR markers were scored as present (1) if the resistance allele was present or (0) if it was absent. Using the present and absent scores for each marker, the Polymorphism Information Content (PIC) values were estimated. The five SSR markers had PIC values of 0.14, 0.36, 0.27, 0.32 and 0.33 for Xpsp3000, Xbarc8, Xwmc44, Xbarc32, and Xgwm419, respectively. Out of the 233 accessions used, all resistance alleles were present in only 11 accessions, which were Sids14, Giza171, “PI 105099”, “PI 14352”, “PI 14284”, “PI 14282”, “PI 14249”, “PI 12811”, “PI 12782”, “PI 12171”, and “PI 11254”. Sids13 had three resistance alleles, while the missing resistance allele was for Xwmc44 and Xgwm419. Gimmiza9 had three resistance alleles, and the missing resistance alleles were Xbarc8 and Xgwm419. Misr2 had two resistant alleles while three alleles were missing, which were Xbarc8, Xwmc44, and Xbarc32. Similarly, Misr1 had two resistance alleles and was missing Xbarc8, Xwmc44, and Xgwm419. Overall, four accessions had none of the resistance alleles investigated, while 26, 120, 40, and 32 accessions contained 1, 2, 3, and 4 resistant alleles, respectively.
A significant positive correlation (p-value=0.001) was detected between grain yield and the five SSR markers used (Table 3). On the other hand, a significant negative correlation was detected between stripe rust scores and the five SSR markers. Moreover, grain yield was negatively correlated with stripe rust (r=-0.87, p-value =0.001). The correlation among the studied SSR markers indicated a significant positive correlation between Xgwm419 and Xpsp3000 markers. Additionally, a significant negative correlation between Xwmc44 and Xbarc32 markers was detected. While the correlation among the other SSR markers were not significant.
Cluster analysis results from the phenotypic measurements (grain yield and stripe rust resistance scores) and SSR markers indicated that the 233 accessions are grouped into two main clusters: A and B (Figure 1). Cluster A contains mainly genotypes with more than two SSR resistance alleles. Cluster B contains accessions with two or fewer resistance alleles. In Cluster A, all accessions with five resistance alleles were grouped in a single subgroup (Figure 1). Interestingly the clustering methodology did not show a clear distinction between the improved accessions and landraces.
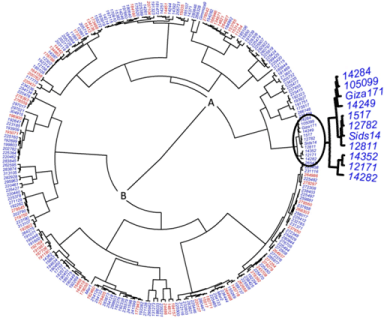
Figure 1: Dendrogram showing the grouping of the 233 wheat accessions, blue and red colors refer to the improved genotypes and landraces, respectively, based
on five SSR markers linked with stripe rust resistance genes, grain yield, and stripe rust scores measured in the field. Magnified accessions are those with five
resistance genes.
The impact of resistance alleles on grain yield and stripe rust resistance was estimated. Accessions with the same number of resistant alleles were plotted together in a boxplot (Figure 1).
The patterns of impact for the number of resistant alleles, from SSR markers, on grain yield production (Figure 2A) and stripe rust resistance (Figure 2B) were similar but with opposite trends. That observed opposite trend was expected because grain yield was negatively correlated with stripe rust resistance scores. As the number of resistance alleles increases, grain yield increases, and stripe rust score decreases (Figure 1). However, the incremental rate between 0 resistance alleles and single resistant alleles was not notable. The boxplot of the accessions with two resistant alleles tended to be divided into two groups, i.e., above-average and below-average groups. The previous observation indicates that the two resistance alleles combinations with different genetic backgrounds (i.e., accessions) resulted in varying grain yield impact levels. In addition, accessions with three resistance alleles produced greater grain yield than those with two resistance alleles. The grain yield and resistant increment rates from 3 to 5 alleles were slower than that from 2 to 3 alleles (Figure 2A and 2B).
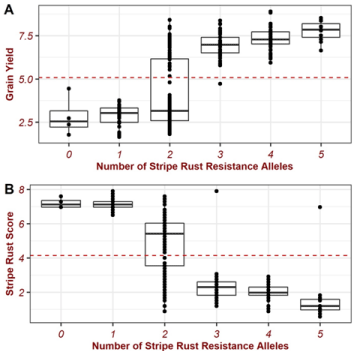
Figure 2: The relationship between the number of stripe rust resistance alleles, grain yield production (A), and stripe rust overall scores under the open field
conditions.
Discussion
Enhancing grain yield production is the goal of plant breeders. However, both biotic and abiotic stresses cause yield loss and endanger the overall grain production worldwide [32]. Among several biotic stresses, recent studies revealed severe impacts of stripe rust on wheat grain production. New stripe rust races have emerged and increased stripe rust susceptibility under field conditions leading to yield losses [33]. The detection of the new races indicates continual changes in virulent stripe rust. Due to these new races, some wheat cultivars previously known to be resistant have become susceptible [33]. Therefore, enhancing commercially grown wheat cultivars with new functional resistance genes is a prerequisite for sustainable wheat production [34]. Continuous screening of the newly introduced wheat accessions under the breeders’ targeted environment to identify potentially resistant genotypes is the most economical and effective strategy to defeat the new rust races and reduce yield losses [35]. In the current study, 278 imported wheat accessions and five commercially grown local wheat cultivars were evaluated under the open field conditions. The evaluation was conducted at Elbostan region in Egypt, one of the world’s hotspots for stripe rust, and where wheat is the dominant winter sown field crop. Our ultimate goals were to identify potentially resistant wheat accessions and investigate the importance of five SSR tagged stripe rust genes on grain yield and stripe rust resistance.
Artificial infection with naturally present stripe rust races, i.e., 0E0, 6E4, 70E20, 128E28, and 134E244, was conducted in Elbostan region. Artificial inoculation is required to ensure adequate levels of infection to differentiate among accessions with varying levels of resistance to all of the primary races of stripe rust. In addition to providing sufficient infection, artificial inoculation also ensures consistent infection types across growing seasons as the climate conditions may change the race mixtures from year to year [7]. Results indicated a highly significant difference among the studied accessions for grain yield and stripe rust resistance. This showed that the accessions varied in response to the stripe rust infection, which was also reflected in grain yield. That causality effect between yield reduction and stripe rust resistance was highly pronounced by the negative and highly significant correlation between both traits, as expected. The negative impact of stripe rust on grain yield production was reported by several authors [1,7-9,12,13,33,36,37]. That negative impact could be attributed to the photosynthesis impairment and assimilates reduction due to stripe rust infection, which reduces flower development, fertilization, and the development of ovules and consequently seed setting [38,39].
Our results also show that the older local cultivars used in the current study tend to be more often infested by stripe rust. For instance, Giza171 [40] and Sids14 [41] were among the newest releases of the Agricultural Research Center (ARC) in Egypt and are also among the most stripe rust resistant accessions in the current study. These findings indicated that the ARC’s wheat breeding programs were successfully incorporating new resistance genes into their elite lines. In addition, these findings also explain the replacement of several older cultivars in Egypt by Giza171 and Sids14 recently. Giza171 was previously found to be resistant to stripe rust. However, the most recent reports from field evaluation in Egypt indicated that Giza171 has become susceptible under field conditions to a newly discovered stripe rust race [8]. Shahin [8] found a new race in several regions in Egypt; however, the closest location he collected samples from was Sakha region, which is around 100km away from Elbostan. Sids14 cultivar was also found to be resistant to stripe rust in the current study; however, to date, no other published reports were found to describe Sids14 resistance to stripe rust under the field conditions in Egypt.
Overall, the top 10 high-yielding and stripe rust resistant accessions originate from improved genotypes, breeders’ elite lines, or registered cultivars. Moreover, the improved genotypes had 11.5, and 11.35% more grain yield and resistance to stripe rust than the landraces. The observed improvement in grain yield production and stripe rust resistance in modern accessions illustrates that several genes for stripe rust were already pyramided. Several genes govern stripe rust-resistant during the seedling and adult plant stages. Five SSR markers were used in the current study to tag and characterize the studied panel for five of the vital stripe rust resistance genes, i.e., Yr10, Yr15, Yr26, Yr29, and Yr59.
The seedling resistance gene Yr10 located on the short arm of chromosome 1B, was tagged in the current study by Xpsp3000 marker, in which it was present in 85.1 and 89.6% of the improved accessions and landraces, respectively. Xpsp3000 was used as a diagnostic marker in MAS for Yr10 and found to be effective in tagging Yr10 [42]. It was previously reported that several of the landraces and improved wheat accessions resistant to stripe rust contained Yr10 [42-44]. Several researchers used the Yr10 gene to provide effective resistance to stripe rust in wheat in most wheat-growing areas; however, recently, several Yr10 virulent Pst races have been reported [43]. Yr15 also had a seedling resistance gene located on the 1B chromosome and was tagged by Xbarc8 and found to be present only in the improved accessions by 23.2%, while none of the landraces had that gene. Yr15 confers broad-spectrum resistance against a large and genetically diverse number of Pst isolates [45]. However, several Pst races were reported to be virulent on Yr15 in 2000 [46].
Yr26 is another seedling resistance gene located near the centromeric region of chromosome 1B tagged with Xgwm419 SSR marker. Yr26 has been widely used in several wheat breeding programs for developing stripe rust-resistant lines [47]. Yr29 is an adult plant resistance gene [48], and it harbors partial resistance showing a slow rusting phenotype [49]. Furthermore, Yr29 is likely a pleiotropic gene in association with leaf rust gene Lr46 and stem rust gene Sr58, hence providing multiple rust disease resistance [49]. Yr59, located the long arm of chromosome 7B, is an adult plant resistant gene and tagged by Xbarc32 [50]. Yr59 was previously reported in an old landrace from Iraq (PI 178759) and found to effective against several Pst races [51].
Landraces harbor valuable stripe rust resistance genes. Therefore, they may play a critical role as gene donors for the new stripe rust races [51,52]. More than 200 stripe rust-resistant landraces were previously reported [53]. It is important to note that Egyptian cultivars tend to harbor at least two out of the five genes investigated, and some of these genes play essential roles in stripe rust resistance. Additionally, several of the imported lines and landraces were susceptible to stripe rust and had low yield potential; thus they were not suited to modern cultivation conditions. However, it is desirable to use the imported wheat entries to develop and maintain representative collections from wheat landraces because they might be useful in transferring desirable traits, such as stripe rust resistance, from these landraces to new elite lines. Overall, the diagnostic SSR molecular markers used in the current study constitute efficient wheat breeding selection tool to identify genuinely resistant lines.
Conclusion
The emergence of new races of stripe rust requires developing and identifying new resistance gene sources. Intensive screening of large local and imported wheat accessions is key in identifying such new resistance genes. In the current study, we were interested in screening, under the field conditions and using SSR markers, an imported collection of wheat landraces, elite lines, landraces, and five recent releases of the wheat cultivars in Egypt. In addition to Giza71 and Sids14, nine of the imported elite lines were found to possess the five known genes investigated in this study. Results from this study suggest that several of the improved accessions were enhanced for stripe rust resistance in several breeding programs. The identified resistant accessions in the current study could be used as a gene source to enhance stripe rust resistance in wheat breeding programs. SSR platform and SSR markers used in the present study effectively capture a substantial part of the phenotypic variation caused by stripe rust. Thus, these five markers could be used effectively in markerassisted selection for stripe rust.
Funding
This work was partially supported by the Science and Technology Development (STDF), Egypt, grant No. 14935, and the winter wheat breeding program in the University of Nebraska-Lincoln, Lincoln, USA.
Acknowledgment
Most of the research work reported in this manuscript was conducted in the wheat-breeding laboratory, Damanhur University, Egypt; for that, the author thanks the wheat breeding team for all their help and technical support. I thank Mr. Hafiz Mazeek and Mr. Talaat Salah, Damanhur University, Egypt, for their effort during this experiment. I would also like to show my gratitude to graduate and undergraduate students who helped during the field and laboratory work.
References
- Gessese MK. Description of Wheat Rusts and Their Virulence Variations Determined through Annual Pathotype Surveys and Controlled Multi- Pathotype Tests. Adv. Agric. 2019: 1-7.
- Huerta-Espino J, R Singh, LA Crespo-Herrera, HE Villaseñor-Mir, MF Rodriguez-Garcia, et al. Adult Plant Slow Rusting Genes Confer High Levels of Resistance to Rusts in Bread Wheat Cultivars From Mexico. Front. Plant Sci. 2020; 11: 1.
- Khanfari S, M BOULIF and R LAHLALI. Yellow Rust (Puccinia striiformis): a Serious Threat to Wheat Production Worldwide. Not. Sci. Biol. 2018; 10: 410-423.
- Figueroa M, KE Hammond-Kosack, and PS Solomon. A review of wheat diseases-a field perspective. Mol. Plant Pathol. 2018; 19: 1523-1536.
- Ali S, P Gladieux, M Leconte, A Gautier, AF Justesen, et al. Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen Puccinia striiformis f.sp. tritici (BA McDonald, editor). PLoS Pathog. 2014; 10: e1003903.
- Ali S, J Rodriguez-Algaba, T Thach, CK Sørensen, JG Hansen, et al. Yellow rust epidemics worldwide were caused by pathogen races from divergent genetic lineages. Front. Plant Sci. 2017; 8: 1057.
- Chen W, C Wellings, X Chen, Z Kang and T Liu. Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2014; 15: 433- 446.
- Shahin AA. Occurrence of new races and virulence changes of the wheat stripe rust pathogen (Puccinia striiformis f.sp. tritici) in Egypt. Arch. Phytopathol. Plant Prot. 2020; 53: 552-569.
- Shahin A, M Ashmawy, W El-Orabey, and S Esmail. Yield Losses in Wheat Caused by Stripe Rust (Puccinia striiformis) in Egypt. Egypt. Am. J. Life Sci. 2020; 8: 127-134.
- Elbasyoni IS, WM El-Orabey, S Morsy, PS Baenziger, Z Al Ajlouni, et al. Evaluation of a global spring wheat panel for stripe rust: Resistance loci validation and novel resources identification (D. Perovic, editor). 2019; 14: e0222755.
- Collard BCY and DJ Mackill. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008; 363: 557-572.
- Mcintosh RA, Y Yamazaki, J Dubcovsky, J Rogers, C Morris, et al. 12th International Wheat Genetics Symposium. 2013: 8-13.
- Basnet BR, RP Singh, AMH Ibrahim, SA Herrera-Foessel, J Huerta-Espino, et al. Characterization of Yr54 and other genes associated with adult plant resistance to yellow rust and leaf rust in common wheat Quaiu 3. Mol. Breed. 2014; 33: 385-399.
- Wu XL, JW Wang, YK Cheng, XL Ye, W Li, et al. Inheritance and molecular mapping of an all-stage stripe rust resistance gene derived from the Chinese common wheat landrace “yilongtuomai.” J. Hered. 2016; 107: 463-470.
- Hartl L, H Weiss, FJ Zeller and A Jahoor. Use of RFLP markers for the identification of alleles of the Pm3 locus conferring powdery mildew resistance in wheat (Triticum aestivum L.). Theor. Appl. Genet. 1993; 86: 959-963.
- Penner GA, J Clarke, LJ Bezte and D Leisle. Identification of RAPD markers linked to a gene governing cadmium uptake in durum wheat. Genome. 1995; 38: 543-547.
- Peng JH, T Fahima, MS Röder, QY Huang, A Dahan, et al. High-density molecular map of chromosome region harboring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetica. 2000; 109: 199-210.
- Lu J, J Hou, Y Ouyang, H Luo, J Zhao, et al. A direct PCR–based SNP marker–assisted selection system (D-MAS) for different crops. Mol. Breed. 2020; 40: 1-10.
- Tan CT, S Assanga, G Zhang, JC Rudd, SD Haley, et al. Development and validation of kasp markers for wheat streak mosaic virus resistance gene wsm2. Crop Sci. 2017; 57: 340-349.
- He D, J Zhang, X Zhang, S He, D Xie, et al. Development of SSR markers in Paeonia based on De Novo transcriptomic assemblies (T-Y Chiang, editor). PLoS One. 2020; 15: e0227794.
- Tommasini L, N Yahiaoui, P Srichumpa, and B Keller. Development of functional markers specific for seven Pm3 resistance alleles and their validation in the bread wheat gene pool. Theor. Appl. Genet. 2006; 114: 165- 175.
- Peterson RF, AB Campbell and AE Hannah. a diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948; 26c: 496-500.
- Roelfs aP, RP Singh and EE Saari. Rust Diseases of Wheat: Concepts and methods of disease management. CIMMYT. 1992.
- Olivoto T and ADC Lúcio. Metan: An R package for multi-environment trial analysis (S. Jarman, editor). Methods Ecol. Evol. 2020; 11: 783-789.
- Smith JSC, ECL Chin, H Shu, OS Smith, SJ Wall, et al. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): Comparisons with data from RFLPS and pedigree. Theor. Appl. Genet. 1997; 95: 163-173.
- Paradis E, J Claude and K Strimmer. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004; 20: 289-290.
- Milligan GW and MC Cooper. A study of standardization of variables in cluster analysis. J. Classif. 1988; 5: 181-204.
- Ali N, I Hussain, S Ali, NU Khan and I Hussain. Multivariate analysis for various quantitative traits in wheat advanced lines. Saudi J. Biol. Sci. 2020.
- TUKEY JW. Comparing individual means in the analysis of variance. Biometrics. 1949; 5: 99-114.
- Steel RGD and JH Torrie. Principles and Procedures of Statistics: A Biometrical Approach. 2nd Ed. McGraw-Hill Publishing Co., New York. 1980.
- Cochran WG and GM Cox. Experimental designs. 2nd ed. Wiley, New York. 1957.
- Raza A, A Razzaq, SS Mehmood, X Zou, X Zhang, et al. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants. 2019; 8.
- Carmona M, F Sautua, O Pérez-Hérnandez and EM Reis. Role of Fungicide Applications on the Integrated Management of Wheat Stripe Rust. Front. Plant Sci. 2020; 11: 733.
- Ellis JG, ES Lagudah, W Spielmeyer and PN Dodds. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 2014; 5: 641.
- Babu P, DK Baranwal, Harikrishna, D Pal, H Bharti, et al. Application of Genomics Tools in Wheat Breeding to Attain Durable Rust Resistance. Front. Plant Sci. 2020; 11: 1324.
- Ash GJ, and JF Brown. Yield Losses in Wheat Caused by Stripe Rust (Puccinia Striiformis West.) in Northern New South Wales. Aust. J. Exp. Agric. 1990; 30: 103-108.
- Draz IS, MS Abou-Elseoud, AEM Kamara, OAE Alaa-Eldein and AF El- Bebany. Screening of wheat genotypes for leaf rust resistance along with grain yield. Ann. Agric. Sci. 2015; 60: 29-39.
- Turner MK, D Ravetta and D Van Tassel. Effect of Puccinia silphii on yield components and leaf physiology in Silphium integrifolium: Lessons for the domestication of a perennial oilseed crop. Sustain. 2018; 10.
- Zetzsche H, W Friedt and F Ordon. Breeding progress for pathogen resistance is a second major driver for yield increase in German winter wheat at contrasting N levels. Sci. Rep. 2020; 10: 1-17.
- Hammad A, A Ado-Warda and A Al-Amaged. Giza 171 : A New High Yielding Bread Wheat Cultivar. Egypt. J. Plant Breed. 2015; 19: 2063-2079.
- Eissa ST, AS Abd El-Hameed and MM Mohamed. Sids 14 a New Bread Wheat Cultivar One-sixth of 14 new varieties of bread wheat. Egypt. J. Plant Breed. 2017; 21: 1059-1077.
- Bariana HS, GN Brown, NU Ahmed, S Khatkar, RL Conner, et al. Characterisation of Triticum vavilovii-derived stripe rust resistance using genetic, cytogenetic and molecular analyses and its marker-assisted selection. Theor. Appl. Genet. 2002; 104: 315-320.
- Liu W, M Frick, R Huel, CL Nykiforuk, X Wang, et al. The Stripe Rust Resistance Gene Yr10 Encodes an Evolutionary-Conserved and Unique CCNBS- LRR Sequence in Wheat. Mol. Plant. 2014; 7: 1740-1755.
- Cheng B, X Gao, N Cao, Y Ding, Y Gao, et al. Genome-wide association analysis of stripe rust resistance loci in wheat accessions from southwestern China. J. Appl. Genet. 2020; 61: 37-50.
- Gerechter-Amitai ZK, CH van Silfhout, A Grama, and F Kleitman. Yr15 - a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Euphytica. 1989; 43: 187-190.
- Liu T, A Wan, D Liu, and X Chen. Changes of races and virulence genes in Puccinia striiformis f.sp. tritici, the wheat stripe rust pathogen, in the United States from 1968 to 2009. Plant Dis. 2017; 101: 1522-1532.
- De-jun HAN, W Qi-lin, Z Li, WEI Guo-rong, Z Qing-dong, et al. Evaluation of Resistance of Current Wheat Cultivars to Stripe Rust in Northwest China, North China and the Middle and Lower Reaches of Changjiang River Epidemic Area. Sci. Agric. Sin. 2010; 43: 2889-2896.
- Cobo N, H Wanjugi, E Lagudah, and J Dubcovsky. A High-Resolution Map of Wheat QYr.ucw-1BL, an Adult Plant Stripe Rust Resistance Locus in the Same Chromosomal Region as Yr29. Plant Genome. 2019; 12: 180055.
- Tehseen MM, FA Tonk, M Tosun, A Amri, CP Sansaloni, et al. Genomewide association study of resistance to PstS2 and Warrior races of Puccinia striiformis f.sp. tritici (stripe rust) in bread wheat landraces. Plant Genome. 2020.
- Zhou XL, MN Wang, XM Chen, Y Lu, ZS Kang, et al. Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI 178759. Theor. Appl. Genet. 2014; 127: 935-945.
- Sthapit J, M Newcomb, JM Bonman, X Chen, and DR See. Genetic Diversity for Stripe Rust Resistance in Wheat Landraces and Identification of Accessions with Resistance to Stem Rust and Stripe Rust. Crop Sci. 2014; 54: 2131-2139.
- Kertho A, S Mamidi, JM Bonman, PE McClean, and M Acevedo. Genomewide association mapping for resistance to leaf and stripe rust in winter-habit hexaploid wheat landraces (G. Bai, editor). PLoS One. 2015; 10: e0129580.
- Wang M, X Chen, L Xu, P Cheng, and HE Bockelman. Registration of 70 Common Spring Wheat Germplasm Lines Resistant to Stripe Rust. J. Plant Regist. 2012; 6: 104-110.
- Elkot A, and M Abd El-Aziz. Molecular Identification of Some Stem Rust and Yellow Rust Resistance Genes in Egyptian Wheat and Some Exotic Genotypes. Assiut J. Agric. Sci. 2016; 47: 124-135.
- Yaniv E, D Raats, Y Ronin, AB Korol, A Grama, et al. Evaluation of markerassisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol. Breed. 2015; 35: 1-12.
- Fayyaz M, A Shahzad, GM Ali, A ur R Rattu and F Muhammad. Identification of stripe rust (Puccinia striiformis) resistant genes among Pakistani spring wheat by using molecular markers. Int. J. Biosci. 2017; 11: 320-334.
- Pourkhorshid Z. Identification of Wheat Stripe Rust Resistance Genes in Iranian Wheat Cultivars Using Molecular Markers. Annu. Res. Rev. Biol. 2014; 4: 2766-2778.