
Special Article - Abiotic Stress
Ann Agric Crop Sci. 2020; 5(3): 1066.
Microsatellite-Based DNA Fingerprinting and Genetic Analysis of Some Selected Aus Rice (Oryza sativa L.) Genotypes
Hossain MA1, Islam MM2, Emon RM3, Rana MS3*, Hossain MA4*, Uddin MI2, Malek MA3, Khan NA2 and Nuruzzaman M4
1Department of Biotechnology, Bangladesh Agricultural University, Bangladesh
2Biotechnology Division, Bangladesh Institute of Nuclear Agriculture, Bangladesh
3Plant Breeding Division, Bangladesh Institute of Nuclear Agriculture, Bangladesh
4Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Bangladesh
*Corresponding author: Md. Shohel Rana, Plant Breeding Division, Bangladesh Institute of Nuclear Agriculture, Mymensingh-2202, Bangladesh
Mohammad Anwar Hossain, Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Mymensingh-2202, Bangladesh
Received: November 30, 2020; Accepted: December 22, 2020; Published: December 29, 2020
Abstract
The allelic diversity and molecular characterization of 30 Aus genotypes were done through DNA fingerprinting using microsatellite (SSR) markers. All the 30 amplified products have polymorphic bands giving 176 alleles. The number of alleles per locus ranged from 4 (RM536) to 20 (RM209), with an average of 9.4889 alleles across 45 loci. A total of 156 rare alleles were detected on 45 loci, whereas 31 null alleles were detected on 30 loci. The Polymorphism Information Contents (PIC) value lied between 0.5511 (RM134) to 0.9199 (RM209). Most robust marker was found to be RM209 since it provided the highest PIC value (0.9199), followed by RM144 (0.912) and RM153 (0.9070). Pair-wise genetic dissimilarity co-efficient showed that the lowest genetic dissimilarity value (0.11) was found between BRRI dhan42 and BRRI dhan43 and the highest genetic dissimilarity value (1.00) was found among all local landraces. The Unweighted Pair Group Method with Arithmetic mean (UPGMA) dendrogram revealed 6 clusters. Cluster I contain seven local landraces: Tusha, Tapa Sail, Udobali, ZamirSaita, Soda, Soil Bogi and Tarabali. Cluster II contain BR1, BR2, BR3, BR6, BR7, BR8, BR9. Cluster III has BR12, BR15, BR16, BR20, BR21. Tepakain is in cluster IV. Cluster V contains BRRI dhan27, BRRI dhan42, BRRI dhan43, BRRI dhan48, BR24, BR26 and cluster VI with SadaBogi, Usha, Sada Aus and Saita. Most Aus landraces is recognized to have broad genetic base. Thus, these landraces can be used for future breeding program or new genes can be incorporated into the landraces to broaden the genetic base.
Keywords: DNA fingerprinting; Genetic analysis; Landraces; Polymorphism Information Contents (PIC); Aus rice
Introduction
Rice is one of the most important cereal crops which grow in all growing seasons of Bangladesh. It grows in all the three crop growing seasons of the year and occupies about 77% (11.42Mha) of the total cropped area of about 14.94 million hectares [1]. According to the United Nations (UN) estimates, the current world population 6.1 billion is expected to reach 8.0 billion by 2025. Bangladesh is already under pressure both from huge and increasing demands for food, and from problems of agricultural land and water resources depletion. Bangladesh needs to increase the rice yield in order to meet the growing demand for food emanating from population growth.
Large variations in morphological, biochemical traits and DNA polymorphism exist in rice in Asia, its center of origin, with sub-centers of diversity in China and Indian subcontinent where both indica and japonica subspecies have been found to grow with abundant variation [2,3]. There is wide genetic variability available within existing varieties of rice and wild relatives providing wide scope for future crop improvement [4,5]. As the number of rice cultivars increases, the ability to distinguish them on the basis of morphological and biochemical traits becomes more difficult mostly due to genotype-environment interaction. Any developed or derived cultivar requires clarity from its precursor for identity and protection. Both breeders and farmers tend to select among variations in their fields in order to maintain the purity of the varieties or screen for a new type. For the study of genetic diversity, the plant scientists have used generally morphological, physiological as well as molecular characterization of plant. Moreover, in most cases, plant genomes have large amount of repetitive DNA which are not expressed and do not contribute to the physiological or morphological appearance of plants. In the case of very closely related plant varieties, there are very few morphological differences, which as a matter of fact do not represent the true genetic differences at DNA level. So, there is always a need to study polymorphism at DNA level, which can be an indicative of genetic diversity [6]. It is thus apparent that the use of molecular genetic markers would provide one solution to the problem of providing unique DNA profiles for the protection of new rice cultivars. With the development of a wide range of molecular techniques, marker-assisted breeding is now used to enhance traditional breeding programs to improve crops [7]. These include Restriction Fragment Length Polymorphism (RFLP), simple sequence repeats (SSR), random amplification of polymorphic DNA (RAPD) and the Amplified Fragment Length Polymorphism (AFLP). PCR-based markers such as microsatellites are co-dominant, hyper variable, abundant and well distributed throughout the rice genome [8]. Microsatellites have shown great promise in genetic diversity, genome mapping, gene tagging and Marker-Assisted Selection (MAS) because they are technically simple, time saving, highly informative and require small amount of DNA. Abundance of microsatellite markers is now available through the published high-density linkage map [9,10] or public database.
Serial No.
Name of the Genotypes
Acc. No.
Origin
Serial No.
Name of the Genotypes
Acc. No.
Origin
1
Soda
1762
Jessore
16
BR6
MV
BRRI
2
Sada Bogi
1965
Jessore
17
BR7 (BRRI Balam)
MV
BRRI
3
Sail bogi
2077
Dhaka
18
BR8 (Aasa)
MV
BRRI
4
Sada Aus
2135
Dhaka
19
BR9 (Sufala)
MV
BRRI
5
Saita (sada)
3547
Faridpur
20
BR12 (Mayana)
MV
BRRI
6
Tarabali
811
Sylhet
21
BR15 (Mohinye)
MV
BRRI
7
Tepakain
1532
Dinajpur
22
BR16 (Sahya Balam)
MV
BRRI
8
Tapa sail
1752
Khulna
23
BR20 (Nizamy)
MV
BRRI
9
Tusha
4567
Dhaka
24
BR21 (Niamat)
MV
BRRI
10
Usha
4570
Dhaka
25
BR24 (Rahmat)
MV
BRRI
11
Udobali
4572
Dhaka
26
BR26 (Sraboni)
MV
BRRI
12
Zamir Saita
4044
Meharpur
27
BRRI dhan27
MV
BRRI
13
BR1 (chandina)
MV
BRRI
28
BRRI dhan42
MV
BRRI
14
BR2 (Mala)
MV
BRRI
29
BRRI dhan43
MV
BRRI
15
BR3 (Biplob)
MV
BRRI
30
BRRI dhan48
MV
BRRI
Table 1: List of rice genotypes used in the experiment.
Bangladesh has a good source of indigenous rice cultivars comprising Boro, Aman and Aus landraces. Those landraces have good adaptation having poor yield. Actually, cultivation of these landraces was gradually replaced by high yielding varieties during the last 20 years. These landraces adapted in different parts of the country, some of which have very nice quality, fineness, aroma, taste and high protein contents [11]. Indigenous crop landraces were characterized at both molecular and phenotypic levels by many countries. Such types of characterization have been done for keeping the crop identity and searching for new genes for further crop improvement. But information on the genetic diversity of local landraces particularly for Aus rice is very scanty in Bangladesh. Precise information on the extent of genetic diversity among population is crucial in any crop improvement program, as selection of plants based on genetic diversity has become successful in several crops. Thus, the research was undertaken to make a DNA fingerprint and genetic relationship among Aus rice genotypes by analyzing the genetic diversity using SSR markers.
Materials and Methods
Plant materials
Thirty genotypes, including eighteen BRRI released Aus varieties were used in this study (Table 1). The experiment was conducted at the experimental field and laboratory of Biotechnology Division, Bangladesh Institute of Nuclear Agriculture (BINA), Mymensingh.
DNA extraction, purification and confirmation
Genomic DNA was extracted from 21-25 days old leaves using the mini preparation modified Cetyl Trimethyl Ammonium Bromide (CTAB) method. DNA confirmation was done by using 0.8% agarose gel electrophoresis.
Documentation of the DNA samples
After electrophoresis, the gel was taken out carefully from the gel chamber and transferred in a prepared ethidium bromide solution for staining. Staining was done for 20 minutes and then placed on the UV transilluminator in the dark chamber of the Image Documentation System (Uvipro Platinum, EU). The UV light of the system was then switched on. The image was visualized on the monitor and the photograph was saved in the Gel Doc computer.
Parental Survey and Primer Selection
Primers were evaluated based on intensity of bands, consistency within individual, presence of smearing and potentiality for population discrimination. In this experiment forty-five primers viz. RM1, RM283, RM237, RM259, RM431, RM452, RM154, RM327, RM514, RM489, RM85, RM307, RM252, RM119, RM178, RM413, RM169, RM153, RM122, RM161, RM541, RM204, RM217, RM11, RM18, RM134, RM25, RM44, RM 105, RM215, RM219, RM171, RM147, RM484, RM216, RM536, RM209, RM167, RM206, RM286, RM144, RM287, RM20, RM519 and RM277 were used for parental surveys and selected.
Allele scoring
The size (in nucleotide base pairs) of the amplified band for each microsatellite marker was determined based on its migration relative to a molecular weight size marker (20bp DNA Ladder) with the help of Alpha Ease FC 4.0 software.
Analysis of SSR Data
The summary statistics including the number of alleles per locus, major allele frequency, gene diversity and Polymorphism Information Content (PIC) values were determined using POWER MARKER version 3.23 [12], a genetic analysis software. Molecular weights for microsatellite products, in base-pairs, were estimated with Alpha Ease FC4.0 software. The individual fragments were assigned as alleles of the appropriate microsatellite loci. Polymorphism Information Content (PIC) values were described by Anderson (1993) [13] for self-pollinated species.
Name of Primer
Product Size (bp)
Primer Sequence
Repeat Motif
Annealing Temp. (°C)
RM1
113
GCGAAAACACAATGCAAAAA
(GA)26
55
GCGTTGGTTGGACCTGAC
RM283
151
GTCTACATGTACCCTTGTTGGG
(GA)18
55
CGGCATGAGAGTCTGTGATG
RM237
130
CAAATCCCGACTGCTGTCC
(CT)18
55
TGGGAAGAGAGCACTACAGC
RM259
162
TGGAGTTTGAGAGGAGGG
(CT)17
55
CTTGTTGCATGGTGCCATGT
RM431
251
TCCTGCGAACTGAAGAGTTG
(AG)16
55
AGAGCAAAACCCTGGTTCAC
RM452
209
CTGATCGAGAGCGTTAAGGG
(GTC)9
55
GGGATCAAACCACGTTTCTG
RM154
183
ACCCTCTCCGCCTCGCCTCCTC
(GA)21
61
CTCCTCCTCCTGCGACCGCTCC
RM327
213
CTACTCCTCTGTCCCTCCTCTC
(CAT)11(CTT)5
55
CCAGCTAGACACAATCGAGC
RM514
259
AGATTGATCTCCCATTCCCC
(AC)12
55
CACGAGCATATTACTAGTGG
RM489
271
ACTTGAGACGATCGGACACC
(ATA)8
55
TCACCCATGGATGTTGTCAG
RM85
107
CCAAAGATGAAACCTGGATTG
(TGG)5 (TCT)12
55
GCACAAGGTGAGCAGTCC
RM307
174
GTACTACCGACCTACCGTTCAC
(AT)14(GT)21
55
CTGCTATGCATGAACTGCTC
RM252
216
TTCGCTGACGTGATAGGTTG
(CT)19
55
ATGACTTGATCCCGAGAACG
RM119
166
CATCCCCCTGCTGCTGCTGCTG
(GTC)6
67
CGCCGGATGTGTGGGACTAGCG
RM178
117
TCGCGTGAAAGATAAGCGGCGC
(GA)5 (AG)8
67
GATCACCGTTCCCTCCGCCTGC
RM413
79
GGCGATTCTTGGATGAAGAG
(AG)11
55
TCCCCACCAATCTTGTCTTC
RM169
167
TGGCTGGCTCCGTGGGTAGCTG
(GA)12
67
TCCCGTTGCCGTTCATCCCTCC
RM153
201
ACCAACGCCAAAAGCTACTG
(GAA)9
55
TACTCGCCCTGCATGAGC
RM122
227
GAGTCGATGTAATGTCATCAGTGC
(GA)7A (GA)2A (GA)11
55
GAAGGAGGTATCGCTTTGTTGGAC
RM161
187
TGCAGATGAGAAGCGGCGCCTC
(AG)20
61
TGTGTCATCAGACGGCGCTCCG
RM541
158
TATAACCGACCTCAGTGCCC
(TC)16
55
CCTTACTCCCATGCCATGAG
RM204
169
GTGACTGACTTGGTCATAGGG
(CT)44
55
GCTAGCCATGCTCTCGTACC
RM217
133
ATCGCAGCAATGCCTCGT
(CT)20
55
GGGTGTGAACAAAGACAC
Table 2(a): Selected primers, product size, their sequences (forward and reverse), Repeat motifs and annealing temperatures.
Name of Primer
Product Size (bp)
Primer Sequence
Repeat Motif
Annealing Temp. (°C)
RM11
140
TCTCCTCTTCCCCCGATC
(GA)17
55
ATAGCGGGCGAGGCTTAG
RM18
157
TTCCCTCTCATGAGCTCCAT
(GA)4AA(GA)(AG)16
55
GAGTGCCTGGCGCTGTAC
RM134
93
ACAAGGCCGCGAGAGGATTCCG
(CCA)7
55
GCTCTCCGGTGGCTCCGATTGG
RM25
146
GGAAAGAATGATCTTTTCATGG
(GA)18
55
CTACCATCAAAACCAATGTTC
RM44
99
ACGGGCAATCCGAACAACC
(GA)16
55
TCGGGAAAACCTACCCTACC
RM 105
134
GTCGTCGACCCATCGGAGCCAC
(CCT)6
55
TGGTCGAGGTGGGGATCGGGTC
RM215
148
CAAAATGGAGCAGCAAGAGC
(CT)16
55
TGAGCACCTCCTTCTCTGTAG
RM219
202
CGTCGGATGATGTAAAGCCT
(CT)17
55
CATATCGGCATTCGCCTG
RM171
290
AACGCGAGGACACGTACTTAC
(GATG)5
55
ACGAGATACGTACGCCTTTG
RM147
97
TACGGCTTCGGCGGCTGATTCC
(TTCC)5 (GGT)5
55
CCCCCGAATCCCATCGAAACCC
RM484
299
TCTCCCTCCTCACCATTGTC
(AT)9
55
TGCTGCCCTCTCTCTCTCTC
RM216
146
GCATGGCCGATGGTAAAG
(CT)18
55
TGTATAAAACCACACGGCCA
RM536
243
TCTCTCCTCTTGTTTGGCTC
(CT)16
55
ACACACCAACACGACCACAC
RM209
134
ATATGAGTTGCTGTCGTGCG
(CT)18
55
CAACTTGCATCCTCCCCTCC
RM167
128
GATCCAGCGTGAGGAACACGT
(CT)18
55
AGTCCGACCACAAGGTGCGTTGTC
RM206
147
CCCATGCGTTTAACTATTCT
(CT)21
55
CGTTCCATCGATCCGTATGG
RM286
110
GGCTTCATCTTTGGCGAC
(GA)16
55
CCGGATTCACGAGATAAACTC
RM144
237
TGCCCTGGCGCAAATTTGATCC
(ATT)11
55
GCTAGAGGAGATCAGATGGTAGTGCATG
RM287
118
TTCCCTGTTAAGAGAGAAATC
(GA)21
55
GTGTATTTGGTGAAAGCAAC
RM20
234
ATCTTGTCCCTGCAGGTCAT
(ATT)14
55
GAAACAGAGGCACATTTCATTG
RM519
122
AGAGAGCCCCTAAATTTCCG
(AAG)8
55
AGGTACGCTCACCTGTGGAC
RM277
124
CGGTCAAATCATCACCTGAC
(GA)11
55
CAAGGCTTGCAAGGGAAG
Table 2(b): Selected primers, product size, their sequences (forward and reverse), repeat motifs and annealing temperatures.
Results and Discussion
The analysis of genetic diversity is very important factor for rice improvement that can be obtained through DNA fingerprinting techniques, which is capable of exhibiting large number of loci for extensive variability.
Forty-five SSR primers (Table 2a&b) were used to analyze the DNA fingerprint and genetic relationship in 30 genotypes of rice this study. In earlier studies, Chakravarthi and Naravaneni, [4] used 30, Islam et al., [14] used 85 and Ahmed et. al., [15] used 12 microsatellite makers as subset for genetic diversity analysis of O. sativa.
In this study, a total of 176 alleles of 45 SSR markers were detected among the 30 rice genotypes. The average number of alleles per locus was 9.4889 with a range of 4 (RM536) to 20 (RM209) (Table 2b). Similarly, Ahmed et al., [15] found a range of 3 alleles (RM234, RM277) to 15 alleles (RM493) with an average of 8.91 and Siddique et. al., [16] found 3 alleles (RM118) to 18 alleles (RM44) with an average of 9.88. Thomson et al., [17] also found the similar range of total number allele in a study from 4 alleles (RM484) to 31 alleles (RM474), with an average of 13.0 alleles across the 30 loci. Comparing microsatellite markers with the different repeat motifs, CT repeats (Table 2) had the largest number of alleles (20). These results of the study reflected that the primers RM209 (20), RM144 (17), RM307 (15), RM (15) are ideal for distinguishing the 30 genotypes as they generate more alleles (Table 2b). Yang et al., [18] also found up to 25 alleles for 10 microsatellite markers among 238 accessions of Indica and Japonica cultivars and landraces.
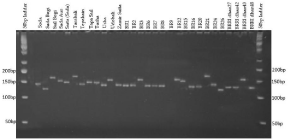
Figure 1: SSR profile of 30 rice genotypes using primer RM209.
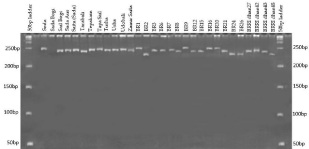
Figure 2: SSR profile of 30 rice genotypes using primer RM536.
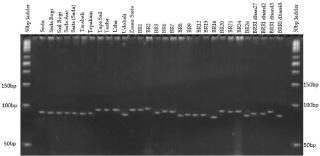
Figure 3: SSR profile of 30 rice genotypes using primer RM489.
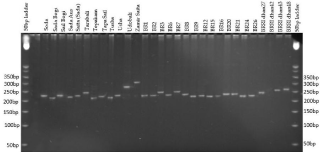
Figure 4: SSR profile of 30 rice genotypes using primer RM134.
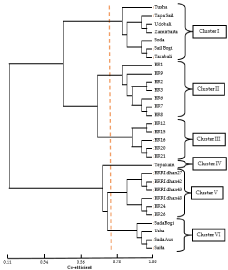
Figure 5: UPGMA dendrogram based on Nei’s (1983) genetic distance
differentiation among 30 rice genotypes. Arrow line indicates the scale of
genetic distance (0.11-1.00).
Locus
Chr. No.
No. of Alleles
Allele Ranges
Difference
Rare Alleles
Null Alleles
PIC
RM1
1
9
80-115
35
2
0
0.8441
RM283
1
8
147-170
23
4
0
0.71
RM237
1
8
126-150
24
3
1
0.7823
RM259
1
12
152-175
23
3
0
0.8765
RM431
1
11
249-270
21
4
1
0.8156
RM452
2
8
190-217
27
1
1
0.8125
RM154
2
10
160-195
35
4
1
0.8019
RM327
2
11
202-218
16
5
1
0.8382
RM514
3
9
245-266
21
1
1
0.837
RM489
3
10
237-311
74
8
1
0.666
RM85
3
9
89-117
28
2
1
0.8177
RM307
4
15
119-173
54
7
1
0.8995
RM252
4
13
196-245
49
6
0
0.8694
RM119
4
7
160-173
13
2
1
0.7838
RM178
5
5
115-124
9
1
1
0.6047
RM413
5
11
71-101
30
4
0
0.8437
RM169
5
8
163-204
41
5
0
0.7195
RM153
5
15
177-203
26
6
1
0.907
RM122
5
8
211-238
27
1
0
0.8119
RM161
5
7
165-186
21
2
0
0.727
RM541
6
8
165-176
11
1
1
0.8202
RM204
6
8
106-144
38
3
1
0.7133
RM217
6
6
124-140
16
0
0
0.777
RM11
7
7
135-147
12
1
0
0.7222
RM18
7
9
153-171
18
4
1
0.7967
RM134
7
6
82-94
8
2
0
0.5511
RM25
8
8
128-158
30
2
1
0.7736
RM44
8
8
104-114
10
0
0
0.8388
RM105
9
11
130-142
12
4
1
0.8348
RM215
9
9
142-154
12
3
1
0.8359
RM219
9
9
195-224
29
2
0
0.7857
RM171
10
13
300-334
34
7
1
0.8637
RM147
10
6
93-99
6
1
1
0.736
RM484
10
5
293-319
26
0
1
0.6957
RM216
10
7
127-147
20
2
1
0.7063
RM536
11
4
238-250
12
0
1
0.5832
RM209
11
20
124-160
36
15
1
0.9199
RM167
11
11
121-159
38
4
1
0.8213
RM206
11
10
128-164
36
2
1
0.8381
RM286
11
14
96-129
33
6
1
0.871
RM144
11
17
214-256
42
11
1
0.912
RM287
11
10
96-119
23
4
1
0.8423
RM20
12
7
155-191
36
2
0
0.6841
RM519
12
15
116-149
33
9
0
0.8919
RM277
12
5
120-127
7
0
1
0.6121
Mean
-
9.489
-
26.1111
3.467
0.667
0.787
Table 3: Data on chromosome numbers, number of alleles, allele ranges, difference of allele ranges, number of rare alleles, number of null alleles and Polymorphism Information Content (PIC) found among 30 rice genotypes for 45 microsatellite (SSR) markers.
Locus
Sample Size
Major Allele
Availability
Gene Diversity
Heterozygosity
Size (bp)
Frequency (%)
RM1
30
114
20
1
0.86
0
RM283
30
147
40
1
0.7444
0
RM237
30
140
26.67
1
0.8089
0
RM259
30
162
20
1
0.8867
0
RM431
30
262
33.33
1
0.8311
0
RM452
30
214
23.33
1
0.8333
0
RM154
30
190
30
1
0.8222
0
RM327
30
202
26.67
1
0.8533
0
RM514
30
245
23.33
1
0.8533
0
RM489
30
248
46.67
1
0.7022
0
RM85
30
97
30
1
0.8356
0
RM307
30
134
16.67
1
0.9067
0
RM252
30
203
23.33
1
0.88
0
RM119
30
161
23.33
1
0.8111
0
RM178
30
124
53.33
1
0.6467
0
RM413
30
101
26.67
1
0.8578
0
RM169
30
204
33.33
1
0.7578
0
RM153
30
201
16.67
1
0.9133
0
RM122
30
237
30
1
0.8311
0
RM161
30
173
40
1
0.7578
0
RM541
30
168
23.33
1
0.84
0
RM204
30
111
40
1
0.7467
0
RM217
30
134
30
1
0.8044
0
RM11
30
124
43.33
1
0.7489
0
RM18
30
160
26.67
1
0.82
0
RM134
30
88
60
1
0.5889
0
RM25
30
150
30
1
0.8
0
RM44
30
106
20
1
0.8556
0
RM105
30
138
30
1
0.8489
0
RM215
30
144
20
1
0.8533
0
RM219
30
204
30
1
0.8089
0
RM171
30
333
20
1
0.8756
0
RM147
30
96
33.33
1
0.7711
0
RM484
30
293
36.67
1
0.7378
0
RM216
30
128
43.33
1
0.7378
0
RM536
30
250
53.33
1
0.6333
0
RM209
30
158
16.67
1
0.9244
0
RM167
30
159
30
1
0.8378
0
RM206
30
161
26.67
1
0.8533
0
RM286
30
99
26.67
1
0.88
0
RM144
30
228
13.33
1
0.9178
0
RM287
30
119
23.33
1
0.8578
0
RM20
30
191
43.33
1
0.7222
0
RM519
30
131
16.67
1
0.9
0
RM277
30
120
50
1
0.66
0
Mean
30
167.822
30.4422
1
0.8093
0
Table 4: Data on sample size, major alleles, availability, gene diversity and heterozygosity found among 30 rice genotypes for 45 microsatellite (SSR) markers.
OTU
1
10
11
12
13
14
15
16
17
18
19
2
20
21
22
23
24
25
26
27
28
29
3
30
4
5
6
7
8
9
1
0
10
0.8
0
11
0.71
0.91
0
12
0.78
0.89
0.56
0
13
0.89
0.91
0.82
0.82
0
14
0.98
0.89
0.89
0.87
0.58
0
15
0.96
0.98
0.93
0.84
0.67
0.42
0
16
0.87
0.89
0.84
0.82
0.6
0.58
0.53
0
17
0.93
0.91
0.89
0.89
0.69
0.6
0.58
0.49
0
18
0.93
0.89
0.84
0.84
0.73
0.53
0.58
0.56
0.4
0
19
0.89
0.93
0.91
0.89
0.67
0.73
0.53
0.53
0.6
0.6
0
2
0.93
0.87
0.96
0.96
0.91
0.93
0.98
0.93
0.93
0.91
0.98
0
20
0.91
0.87
0.89
0.91
0.69
0.71
0.69
0.6
0.58
0.58
0.64
0.89
0
21
0.87
0.87
0.93
0.87
0.78
0.8
0.78
0.69
0.67
0.67
0.78
0.87
0.58
0
22
0.8
0.91
0.84
0.93
0.8
0.84
0.82
0.69
0.69
0.62
0.82
0.87
0.69
0.58
0
23
0.93
0.91
0.82
0.91
0.8
0.8
0.82
0.76
0.69
0.78
0.78
0.93
0.67
0.62
0.62
0
24
0.87
0.96
0.89
0.89
0.82
0.82
0.78
0.78
0.78
0.8
0.78
0.91
0.71
0.73
0.58
0.53
0
25
0.98
0.93
0.96
0.98
0.82
0.93
0.93
0.82
0.89
0.87
0.89
0.84
0.93
0.91
0.87
0.91
0.91
0
26
1
0.96
0.93
0.96
0.87
0.98
0.96
0.89
0.93
0.93
0.93
0.84
0.93
0.93
0.89
0.89
0.87
0.27
0
27
0.98
0.89
0.93
0.98
0.91
0.93
0.91
0.87
0.93
0.89
0.93
0.84
0.84
0.91
0.82
0.87
0.82
0.44
0.44
0
28
0.98
0.96
0.89
0.96
0.91
0.91
0.91
0.93
0.96
0.89
0.96
0.87
0.89
0.96
0.87
0.89
0.82
0.47
0.49
0.4
0
29
0.98
0.98
0.89
0.96
0.93
0.93
0.93
0.93
0.96
0.91
0.98
0.84
0.91
0.96
0.87
0.89
0.84
0.47
0.49
0.47
0.11
0
3
0.58
0.82
0.71
0.76
0.82
0.87
0.91
0.87
0.89
0.89
0.84
0.93
0.93
0.89
0.89
0.84
0.87
0.98
0.98
1
0.98
0.96
0
30
1
0.96
0.93
0.93
0.91
0.93
0.91
0.89
0.91
0.91
0.96
0.8
0.93
0.93
0.89
0.87
0.87
0.44
0.42
0.53
0.47
0.4
0.98
0
4
0.91
0.78
0.89
0.91
0.89
0.89
0.98
0.93
0.93
0.93
0.96
0.76
0.87
0.96
0.93
0.89
0.93
0.82
0.8
0.84
0.84
0.87
0.91
0.82
0
5
0.87
0.67
0.87
0.93
0.89
0.91
0.98
0.91
0.91
0.93
0.98
0.84
0.87
0.91
0.91
0.89
0.96
0.87
0.84
0.78
0.89
0.93
0.93
0.89
0.56
0
6
0.69
0.82
0.64
0.71
0.87
0.91
0.91
0.87
0.87
0.91
0.87
0.93
0.89
0.87
0.84
0.84
0.84
0.96
0.93
0.96
0.96
0.96
0.49
0.96
0.89
0.87
0
7
0.96
0.87
0.93
0.91
0.93
0.93
0.93
0.96
0.87
0.91
0.91
0.84
0.89
0.91
0.87
0.89
0.89
0.93
0.91
0.96
0.96
0.96
0.96
0.91
0.91
0.89
0.93
0
8
0.69
0.84
0.62
0.6
0.82
0.89
0.93
0.84
0.84
0.8
0.84
0.91
0.82
0.84
0.8
0.87
0.84
0.96
0.93
0.96
0.96
0.96
0.6
0.96
0.89
0.89
0.64
0.91
0
9
0.73
0.91
0.71
0.76
0.82
0.8
0.84
0.84
0.82
0.82
0.8
0.98
0.78
0.8
0.91
0.82
0.84
0.96
0.93
0.96
0.91
0.91
0.69
0.93
0.93
0.96
0.67
0.91
0.71
0
Table 5: Nei's (1983) coefficient of genetic distance for SSR markers.
An allele observed in less than 5% of the 30 rice genotypes were considered to be rare. Rare alleles were observed at all of the SSR loci with an average of 3.467 rare alleles per locus and a total of 156 across all the loci (Table 3). Similarly, Ahmed et. al., [15] observed average of 3.63 rare alleles per locus and a total of 40 across all the loci. This result shows some dissimilarities with the average of 7 rare alleles [19]. In general, markers detecting a greater number of alleles per locus detected rarer alleles. Marker RM209 detected the highest number of alleles (20) and rare alleles (15) simultaneously. Totally absent of allele indicates null allele. In this experiment the average value of null allele is 0.6667. Among the 45 markers, 30 markers have one (1) null allele and the rest 15 markers have none (Table 3).
Polymorphism Information Content value reflects allele diversity and frequency among varieties. PIC value of each marker can be evaluated on the basis of its alleles. According to the measure of the informative nature of microsatellites, the PIC values ranged from 0.5511 (RM134) to 0.9199 (RM209) with a mean PIC value of 0.7866 (Table 3). The PIC values observed, are comparable to two previous estimates of microsatellite analysis in rice viz., 0.65- 0.91 [20], 0.59-0.90 [21], 0.08-.90 with an average 0.69 [15]. The higher PIC values in RM209 and RM144 revealed that RM209 and RM144 are the best markers for distinguishing 30 Aus landraces. PIC values also showed a significant, positive correlation with the number of alleles and allele size range for microsatellites evaluated in this study. However, lower PIC values were found in rest of the markers which may be the result of closely related genotypes and higher PIC values might be the result of diverse genotypes [22]. The number of alleles amplified by a primer and its PIC values also depends upon the repeat number and the repeat sequence of the microsatellite sequences [8,23].
Major allele is defined as the allele with the highest frequency and also known as most common allele at each locus. The size of the different major alleles at different loci ranges from 111bp (RM204) to 333bp (RM171) (Table 3). On average, 30.44% of the 30 rice genotypes shared a common major allele ranging from 40% (RM204) to 20% (RM171) common allele at each locus. The observations show similarity with some previous studies [4,15,21].
According to Nei’s [24], the highest level of gene diversity value (0.9244) was observed in loci RM209 and the lowest level of gene diversity value (0.6333) was observed in loci RM536 with a mean diversity of 0.8093 (Table 4). It was observed that marker detecting the lower number of alleles showed lower gene diversity than those, which detected higher number of alleles, which revealed higher gene diversity. The maximum number of repeats within the SSRs was also positively correlated with the genetic diversity.
Genetic similarities were calculated from the data of Nei’s [24] coefficient (Table 5). The similarly matrix was used to determine the level of relatedness among the studied genotypes. Pair-wise estimates of similarity ranged from 0.11 to 1.00 with other similarities of 0.27, 0.40, 0.42, 0.44, 0.47, 0.49, 0.53, 0.56, 0.58, 0.60, 0.62, 0.6444, 0.67, 0.69, 0.71, 0.73, 0.76, 0.78, 0.80, 0.82, 0.84, 0.87, 0.89, 0.91, 0.93, 0.96 and 0.98. The lowest genetic distance (0.11) was observed in BRRI dhan42 vs. BRRI dhan43. The highest genetic distance of 1.00 was observed between three pairs: Soda vs. BRRI dhan26, Soda vs. BRRI dhan48 and in Sail Bogi vs. BRRI dhan27. Similar results were observed by Ahmed et al., [15] but Hossain et al., [25] observed highest similarity of 0.81 and lowest of 0.21, which is slightly different with this study.
Dendrogram based on Nei’s [24] genetic distance using Unweighted Pair Group Method of Arithmetic Means (UPGMA) indicated differentiation of the 30 rice genotypes (by 45 markers). All 30 rice genotypes could be easily distinguished. The dendrogram obtained using the UPGMA method revealed cultivars that were genetically similar and thus clustered together and explained the relationship among the test rice varieties. The UPGMA cluster analysis led to the grouping of the 30 genotypes in six major clusters at genetic similarity level of 0.71 to 0.82 (Figure 5). Earlier observation also showed six clusters at genetic similarity level of 0.43 to -0.58 [15] and 0.40 [25]. The above results suggest that the local varieties in cluster I share common ancestors whereas, the local varieties in cluster VI are close relatives. It shows the modern varieties BR1, BR2, BR3, BR6, BR7, BR8, BR9, BR12, BR15, BR16, BR20 and BR21 are closely related to cluster I varieties at co-efficient 0.45. On the other hand, BR24, BR26, BRRI dhan27, BRRI dhan42, BRRI dhan43, BRRI dhan48 show relation to the local variety Tepakain at co-efficient 0.67 which suggests they have been developed from Tepakain. Interestingly, all the high yielding rice varieties clustered in the same group and BRRI released varieties clustered in the same group, showed the maximum divergence from the other clusters. In a previous study, 29 rice genotypes were reported to have been grouped into two distinct clusters with the aid of 20 SSR markers [26].
Conclusion
Genetic information gathered here provides unique DNA profiles for Bangladeshi Aus landraces, which will serve as a strong weapon to protect our breeders IPR. Moreover, these data will help the breeders to select parents for future breeding programs as the identification and utilization of diverse genetic resources is a prerequisite for plant improvement. Here it is found that most of the Aus landraces is recognized to have broad genetic base. Thus, it is recommended to use these landraces for future breeding program to include new and untouched landraces in order to incorporate new genes for broadening genetic base.
Acknowledgement
The authors would like to thank the University Grants Commission and Bangladesh Agricultural University for providing the research grant. Special thanks to Bangladesh Institute of Nuclear Agriculture for providing the research facilities.
References
- BBS 2012. Bangladesh Bureau of Statistics. Statistical, Year Book of Bangladesh, Ministry of Planning. Government of the People's Republic of Bangladesh. Dhaka.
- Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theoretical and Applied Genetics. 2001: 95: 553-567.
- Noldin JA. Red rice status and management in Americas. Baki BB, editors. In: Wild and weedy rice in rice ecosystems in Asia: a review. International Rice Research Institute. Los Banos. 2000; 21-24.
- Chakravarthi BK, Naravaneni R. SSR marker-based DNA fingerprinting and diversity study in rice (Oryza sativa L). African Journal of Biotechnology. 2006; 5: 684-688.
- Rahman MS, Easmin F, Islam MS, Samad MA, Alam MS. Random Amplified Polymorphic DNA (RAPD) analysis in some indigenous aromatic rice (Oryza sativa L.) cultivars. Bangladesh Journal of Crop Science. 2007; 18: 331-340.
- Sunnucks P. Efficient genetic markers for population biology. Tree. 2001; 15: 199-203.
- Collard BCY, Mackil DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Transactions of the Royal Society of London. 2008; 363: 557-572.
- Temnykh S, Clerck WD, Carthinour S, Lukashova A, Lipovich L, McCouch SR. Computational and experimental analysis microsatellites in rice (Oryza sativa L.): frequency, length variation, transpose associations and genetic marker potential. Genome Research. 2011; 11: 1441-1552.
- IRGSP (International Rice Genome Sequencing Project). The map-based sequence of the rice genome. Nature. 2005; 436: 793-800.
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Research. 2002; 9: 199-207.
- Dutta RK, Lahiri BP, Mian MAB. Characterization of some aromatic and fine rice cultivars in relation to their phsico-chemical quality of grains. Indian Journal of Plant Physiology. 1998; 3: 61-64.
- Liu K, Muse SV. Power Marker: Integrated analysis environment for genetic marker data. Bioinformatics. 2005; 21: 2128-2129.
- Anderson JA, Churchill, GA, Autrique JE, Tanksley SD, Sorrells V. Optimizing parental selection for genetic linkage maps. Genome. 1993; 36: 81-186.
- Islam MM, Begum HA, Ali MS, Kamruzzaman M, Hoque S, Hoque MI. DNA fingerprinting and genetic diversities in some Bangladeshi Aus rice (Oryza sativa L.) genotypes. SAARC Journal of Agriculture. 2017; 15: 123-137.
- Ahmed R, Islam MM, Emon RM, Rana MS, Haque MS, Nuruzzaman M. DNA Fingerprinting and diversity analysis of some Aus rice landraces. Indian Journal of Science and Technology. 2019; 12.
- Siddique MA, Khalequzzaman M, Fatema K, Islam MZ, Baktiar MHK, Islam MM. DNA Fingerprinting and genetic diversity in Aus rice (Oryza sativa L.) landraces of Bangladesh. Bangladesh Rice Journal. 2017; 21: 59-65.
- Thomson MJ, Septiningsih EM, Suwardjo F, Santoso TJ, Silitonga TS, McCouch SR. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theoretical and Applied Genetics. 2007; 114: 559-568.
- Yang GP, Maroof M, Xu C, Zhang Q, Biyashev R. Comparative analysis of microsatellite DNA polymorphism inland races and cultivars of rice. Molecular and General Genetics. 1994; 245: 187-194.
- Hassan MM, Shamsuddin AKM, Islam MM, Khatun K, Halder J. Analysis of genetic diversity and population structure of some Bangladeshi rice landraces and HYV. Journal of Science Research. 2012: 4: 757-767.
- Siddique MA, Rashid ESMH, Khalequzzaman M, Bashar MK, Khan LR. Molecular characterization and genetic diversity in T Aman landraces of Rice (Oryza sativa L.) using microsatellite markers. Thai Journal of Agricultural Science. 2014; 47: 211-220.
- Siddique MA, Khalequzzaman M, Islam MM, Fatema K, Latif MA. Molecular characterization and genetic diversity in Geographical Indication (GI) rice (Oryza sativa L.) cultivars of Bangladesh. Brazilian Journal of Botany. 2016; 1-10.
- Rahman MS, Sohag MKH, Rahman L. Microsatellite based DNA fingerprinting of 28 local rice (Oryza sativa L.) varieties of Bangladesh. Journal of Bangladesh Agricultural. University. 2010; 8: 7-17.
- Yu SB, Xu WJ, Vijayakumar CHM, Ali J, Fu BY, Xu JL, et al. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theoretical and Applied Genetics. 2003; 108: 131-140.
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences. USA. 1983; 70: 3321-3323.
- Hossain MZ, Rasul MG, Ali MS, Iftekharuddaula KM, Mian MAK. Molecular characterization and genetic diversity in fine grain and aromatic landraces of rice using microsatellite markers. Bangladesh Journal of Plant Breeding and Genetics. 2007; 20: 1-10.
- Upadhyay P, Singh VK, Neeraja C. Identification of genotype specific alleles and molecular diversity assessment of popular rice (Oryza sativa L.) varieties of India. International. Journal of Plant Breeding and Genetics. 2011; 5: 130-140.