
Research Article
Austin Aging Res. 2025; 4(1): 1008.
Exploring Biomarker Correlations in the Aging Process
Naik NN and Suresh P*
Acasta Health Pvt Ltd, India
*Corresponding author: Poosala Suresh, Acasta Health Pvt Ltd, VCR Park, Plot No. 9&12 Layout vide TP No. 9/74, Survey No. 119, Krishna Nagar, Maharani Peta, Visakhapatnam, AP 530002, India Tel: +919100851758; Email: suresh@acastahealth.com
Received: February 20, 2025; Accepted: March 12, 2025; Published: March 17, 2025
Abstract
Aging has traditionally been perceived as an inevitable decline, marked by the coexistence of wisdom and frailty. However, emerging research reveals that aging is not merely an unavoidable process but a condition with identifiable and modifiable characteristics. While debates on a single underlying cause of aging remain unresolved, consensus has been reached on 12 biological hallmarks of aging, each representing a measurable facet of the aging process that informs targeted interventions. This systematic review focuses on the interplay between key genomic and proteomic biomarkers associated with these hallmarks of aging. By analysing biomarker correlations and their implications, the review offers critical insights into mechanisms influencing biological age, health span, and longevity. A comprehensive methodology was employed to identify trends, gaps, and opportunities in biomarker studies. These findings underscore the importance of multi-omics integration in unravelling the complexities of aging. By bridging genomic, proteomic, metabolomic, and functional biomarkers, this approach provides a comprehensive understanding of the aging process. It establishes a foundation for innovative interventions to promote healthy aging, extend health span, and enhance overall quality of life.
Keywords: Aging biomarkers; Proteomics; Genomics; Systems biology; Multi-omics integration; Healthspan; Longevity
Introduction
In recent decades, there has been a steady increase in the average lifespan, given advancements in healthcare and medicine. Thus, the world population is witnessing a shift towards an older population. In biological or physiological terms, aging can be defined as the declining functional capacity of the human body over time, caused by the accumulation of various molecular and cellular damage and increasing loss of cellular and tissue homeostasis [1]. On the other hand, chronological age (CA) is the number of years since an individual has been alive. The chronological age and biological age of an individual might not be the same, as the rate of aging might differ among individuals depending on various internal and external factors. For example, if an individual is healthy and fit, their biological age may well be lower than or same as their chronological age. However, if an individual is sedentary, chronically ill, or in poor physical condition, their biological age may be higher than their chronological age, indicating possible future risk of certain age-related complications.
Moreover, because the aging process is usually slow and gradual, determining biological age and aging rate presents opportunities and options for successful and healthy aging with appropriate guided lifestyle changes [2]. Biological age (BA) conveys the physiological status of your body, which is affected by diet, exercise, lifestyle, comorbidities or predisposition for comorbidities, external environmental stressors, and the natural aging process. It is an indicator of overall health and wellness and of that of various organs. In addition, BA indicates the physiological aging rate of an individual compared to that expected for the corresponding chronological age, thus leading to an altered risk of experiencing age-related complications [1]. BA also acts as a guide to making personalized lifestyle changes to improve overall health and prevent or delay aging-related indications. Biological age (BA) is important for clinical monitoring, community surveillance, and evaluating interventions to delay or prevent agingrelated disorders and disabilities. Clinical and cellular biomarkers can be measured and integrated in years using mathematical models to display an individual’s BA [3]. This disparity between chronological and biological age underscores the importance of understanding the mechanisms behind aging and developing strategies for successful aging.
In recent decades, different aging biomarkers have been studied and explored in population studies to estimate biological age, both of organs and the overall body. The biological age-determining methods/ models based on such studies are often called “age-predictive methods” or commonly referred to as “aging clocks” [1]. A comprehensive review Jylhävä et.al summarized current state-of-the-art findings considering various types of biological age predictors. Jylhävä et.al stated that the existing biological age predictors provide additional evidence on individual aging independent of their chronological age and predict health outcomes such as physical function, cognition, morbidity, and mortality. It is imperative to have a validated set of markers to predict biological age that provides insight into health span rather than only focusing on mortality and lifespan, thus moving the focus towards successful and healthy aging. Ideally, the marker combinations could include a set of physiologic, genomic, and proteomic markers [4].
In line with the same, a varied set of markers predicting biological age is more relevant and accurate since aging is a complex process occurring at all levels of an individual. Biomarkers derived from various hallmarks of aging or ‘mechanistic underpinnings of aging’ present the possibility of measuring the aging processes before clinically recognizable symptoms are visible [5].
Human aging is a complex phenomenon and exploring the intricacies of aging requires a multidisciplinary approach combining biology, genetics, cellular physiology among other fields. Researchers have extensively studied some of the molecular mechanism underlying aging, such as telomere shortening, DNA damage, and oxidative stress and their contribution to the decline in cellular function and the development of age-related diseases [6, 7, 8]. The study of aging, given the complexity of the subject, involves understanding the interplay between genetic factors, environmental influences, and lifestyle choices. Scientific advancements have highlighted potential interventions to support healthy aging, including caloric restriction and pharmaceutical approaches targeting cellular pathways [ 9, 10]. By employing a scientific approach, researchers strive to uncover the mechanisms of human aging, laying the groundwork for strategies to improve well-being and extend health span in an aging population.
This review examines biological aging, a process influenced by genetics, lifestyle, and environmental stressors. Comprehensive assessments of biological age increasingly utilize aging clocks, which integrate data from multiple biomarkers to provide a holistic understanding of the aging process. By synthesizing evidence on key genomic and proteomic biomarkers, this review highlights their interrelationships and relevance in advancing aging research. It also delves into the hallmarks of aging, exploring how each biomarker discussed is intricately connected to these hallmarks. By focusing on a select few genomic and proteomic markers, the review underscores the critical role of biomarkers in unravelling the complexities of aging and provides valuable insights into their applications for aging research and targeted interventions.
Methodology
We conducted a comprehensive search across major scientific databases, including PubMed, Scopus, Web of Science, and Google Scholar. Articles were identified using keywords such as “aging biomarkers,” “biological age,” “genomic biomarkers,” “proteomic biomarkers,” and “multi-omics in aging,” with Boolean operators and truncation applied to refine the search results. The inclusion criteria focused on articles and reviews discussing the roles of genomic and proteomic biomarkers in aging, as well as studies providing insights into biomarker correlations. Literature unrelated to aging biomarkers and non-English publications were excluded.
The findings from the selected studies were narratively organized, with data categorized by biomarker types and analysed for common themes and correlations. This approach highlighted key trends, significant insights, and gaps in the current literature, laying the groundwork for identifying future research directions in aging biomarker studies. A shortlist of genomic and proteomic biomarkers was created based on their association with the hallmarks of aging.
Hallmarks of Aging
Experts have long debated a single underlying cause for aging; instead, they have reached a consensus on multiple biological “hallmarks of aging.” Today, we recognize 12 hallmarks of aging (Figure 1), each representing a measurable aspect of the process that can guide targeted interventions and support healthier, more resilient aging. These hallmarks are interconnected among each other [11]. Initially nine molecular, cellular, and systemic hallmarks of aging: DNA instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication, were suggested in 2013 [12]. A decade later, three additional hallmarks of aging: disabled macro autophagy, chronic inflammation, and dysbiosis were added, and some reorganizations were introduced to the existing nine hallmarks of aging [11].
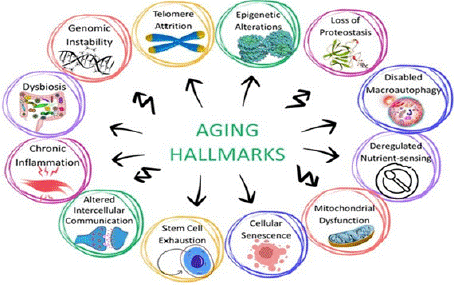
Figure 1: Hallmarks of Aging.
The interconnected nature of aging hallmarks means that experimentally amplifying or reducing one hallmark often influences others. This highlights the complexity of aging as a process that must be understood holistically [11]. To address this complexity, it is crucial to include a diverse range of biomarkers that capture the interplay between these hallmarks, providing a comprehensive understanding of aging.
Types of Aging Biomarkers: Current Understanding
An aging biomarker, individually or combined with other variables, is a physiological measure to detect, diagnose, or forecast the functional competence or function loss of any biological component of a live organism in the absence of illness [13]. Building on this foundational understanding, we now delve into the different categories of aging biomarkers- genomic and proteomic biomarkers- each providing unique insights into various hallmarks of aging. A compilation of genomic and proteomic biomarkers is shown in Table 1. Given the massive scope of the topic, we will limit to a select few biomarkers associated to various hallmark of aging.
Biomarkers
Associated Hallmark of Aging
Telomere Length
Telomere Attrition
AMP-activated protein kinase (AMPK)
Autophagy
Sirtuin 6 (SIRT6)
Mitochondrial Dysfunction & Genomic Instability
Secreted Protein Acidic and Rich in Cysteine (SPARC)
Cell Senescence
4-Hydroxy-2-nonenal (4-HNE)
Inflammation and Cell Senescence
SRY-Box Transcription Factor 2 (SOX2)
Stem cell exhaustion
C-reactive protein (CRP)
Inflammation
Taurine
Increased Cellular Senescence, Telomerase Deficiency, Mitochondrial Dysfunction, DNA Damage, and Inflammation
Interleukin-6
immune-senescence
Mammalian/mechanistic target of rapamycin (mTOR)
Cell senescence/ Mitochondrial dysfunction/Autophagy
DNA (cytosine-5)-methyltransferase 1 (DNMT-1)
Epigenetics/Methylation
Aconitase-2
Mitochondrial dysfunction, Autophagy
Interleukin 8
Cell senescence, Inflammation
Phosphatidylinositol 3-kinase (PI3K)
genome instability, telomere erosion, epigenetic alterations, and mitochondrial dysfunction.
Table 1: Genomic and Proteomic Biomarkers and their associated hallmarks of aging.
Genomic Biomarkers
Based on extensive large-scale studies conducted over several decades, longevity appears to be only moderately heritable. The genetic influences on longevity are likely non-additive, while the environmental factors contributing to it are non-shared [14, 15]. Despite this, the genetic component of lifespan variation across the general population has been estimated to account for approximately 25% [16].
One key area where genetic factors intersect with the biology of aging is the structure and function of telomeres. Telomeres are repetitive DNA sequences (TTAGGG) that protect chromosomes during cell division and are essential for DNA replication. However, mammalian cells experience telomere shortening with each division, limiting their capacity to divide. As a result, cells lose their proliferative potential and enter a state of irreversible cell cycle arrest, known as replicative senescence.
Telomere length (TL) is a critical determinant of cellular replicative capacity, particularly in tissues like the skin, which are highly susceptible to accelerated telomere shortening due to external DNA-damaging agents such as solar radiation, pollution, and reactive oxygen species (ROS) [17]. In multicellular organisms, TLs are highly heterogeneous across tissues and cell types, reflecting differences in tissue-specific proliferation rates. Nevertheless, telomere shortening occurs universally in all proliferating tissues with age [18]. Telomereinduced senescence has been identified as a potential key driver of aging [19].
A large community-based study in Scotland found TL to be significantly associated with age and eight measures of physical and cognitive functioning, as well as overall health status—factors closely tied to normal aging [20]. TL serves as a marker of cellular senescence and chronic disease-related oxidative stress. Accelerated telomere shortening and reduced telomerase activity have been linked to age-related skeletal pathologies like osteoporosis and osteoarthritis, caused by abnormal subchondral bone remodelling [21]. Similarly, shorter TLs elevate the risk of pathologies involving restricted cellular proliferation and tissue degeneration, such as atherosclerosis-related cardiovascular disorders [22].
Both genetic and environmental factors influence TL and the rate of age-related telomere shortening [22]. Developmental experiences, such as unfavorable intrauterine conditions [23], TL at birth [24], and early life adversity (e.g., low socioeconomic status, neglect, or abuse) [25], have profound long-term effects on telomere dynamics. Additionally, adult-life exposures, including infections [26], psychoemotional stress [27, 28], nutrition [29], physical activity [30], smoking [31], and alcohol consumption [32], significantly impact TL and overall cellular aging. While telomeres provide critical insights into cellular senescence and aging, another emerging and equally significant area of research focuses on epigenetic modifications, particularly DNA methylation, which offers a dynamic and reversible layer of regulation influencing the aging process. Recent studies have identified a measure of DNA methylation age, also referred to as the epigenetic clock, as a viable biological age predictor.
Among these, the Horvath (2013) and Hannum (2013) epigenetic clocks are currently considered the most robust and reliable predictors of chronological age. Both show high correlations with age (r = 0.96 for Horvath and r = 0.91 for Hannum) and minimal mean deviations from calendar age (3.6 years and 4.9 years, respectively) in their respective validation cohorts [33, 34]. The Horvath clock was developed using a large sample size of approximately 8,000 individuals spanning the entire adult lifespan and multiple ethnic populations. It is a multitissue predictor that utilizes methylation levels of 353 CpG sites from the Illumina 27k array. In contrast, the Hannum clock, based on 656 individuals, focuses on 71 CpG sites from the Illumina 450k array and is most accurate with whole blood samples [35].
What sets these clocks apart is their ability to predict allcause mortality, independent of traditional risk factors. A metaanalysis across 13 cohorts, comprising a total of 13,089 individuals, demonstrated that the epigenetic clock could predict all-cause mortality independent of factors such as age, BMI, education, smoking, physical activity, alcohol use, and comorbidities [36].
Genomic biomarkers such as telomere length and epigenetic biomarkers not only reveal the cumulative effects of genetic and environmental factors but also underline the dynamic interplay between molecular integrity and aging processes. Furthermore, accumulation of genomic instability from DNA damage caused by endogenous factors (e.g., replication errors, reactive oxygen species) and exogenous stressors (e.g., radiation, toxins), also characterize aging. This genomic instability, which is one of hallmarks of aging, leads to mutations, chromosomal abnormalities, and impaired cellular function, driving aging and age-related diseases.
In addition to genomic factors, the aging process is intricately influenced by changes in the proteome—the complete set of proteins expressed in a cell or organism. Proteins serve as the functional molecules of life, mediating critical biological processes and cellular communication. However, aging disrupts proteostasis, another hallmark of aging, the delicate balance between protein synthesis, folding, and degradation, leading to the accumulation of damaged or misfolded proteins. In the next section, we will explore key proteomic biomarkers and their roles in unravelling the hallmarks of aging, focusing on their significance in maintaining cellular function and systemic health.
Proteomic Biomarkers
Proteomics offers a comprehensive and quantitative view of the entire protein expression landscape within an organism, reflecting the dynamic state of cellular processes and their responses to environmental or biological perturbations [37]. As humans progress through different stages of life—from neonates to adulthood—an array of proteins is differentially expressed, showcasing the profound impact of age on the proteomic profile [38]. Proteomic biomarkers provide real-time insights into the mechanisms underlying the hallmarks of aging and the disruptions associated with age-related processes. Beyond proteostasis, specific proteomic biomarkers can illuminate diverse hallmarks of aging, guiding targeted approaches to address these biological changes. By analyzing protein expression patterns, modifications, and interactions, researchers can identify unique signatures of aging as well as early indicators of age-associated diseases, advancing our understanding and management of the aging process.
A twin-based cohort study identified four replicating proteins that demonstrated significant independent associations with age, highlighting the potential of proteomic biomarkers to serve as robust indicators of the aging process [39]. These findings underscore the value of proteomic studies in identifying proteins that not only reflect biological age but also provide insights into the molecular mechanisms underlying age-related changes. Such proteins could serve as critical tools in monitoring aging trajectories and developing interventions to promote healthy aging.
Building on the foundational understanding of proteomics, we now delve deeper into specific protein biomarkers associated with individual hallmarks of aging. By examining key protein biomarkers associated with various hallmarks of aging, we can gain targeted insights into the molecular disruptions driving age-related changes and identify potential pathways for intervention and therapeutic strategies.
AMP-activated Protein Kinase
One such critical protein biomarker that bridges metabolic regulation and aging is AMP-activated protein kinase (AMPK). This regulator of cellular energy homeostasis plays a pivotal role in modulating various hallmarks of aging, making it a key focus in aging research and therapeutic development. AMPK plays a crucial role in cellular energy regulation by responding to energy stress through phosphorylation, which restores ATP levels by inhibiting energy-consuming pathways and activating ATP-generating catabolic pathways. AMPK also influences longevity and inter-tissue communication, as intestine-specific AMPK upregulation activates autophagy both locally and in distant tissues like the brain, reduces proteotoxicity, and extends lifespan [40]. Autophagy, regulated by AMPK, is vital for degrading and recycling cellular components, maintaining homeostasis, and facilitating inter-tissue communication via the release of cytosolic molecules [41]. Tools such as Western blotting to measure AMPK Thr
(172) phosphorylation and mRNA expression analysis of AMPK subunits are pivotal for studying its pathway [42].
Overall, AMPK-mediated pathways not only maintain cellular energy homeostasis but also modulate aging and tissue health in a non-cell-autonomous manner, underscoring its significance in health and disease. While AMPK plays a central role in maintaining cellular energy homeostasis and autophagy, another key regulator, SIRT6, contributes to preserving mitochondrial function and genomic stability—two critical hallmarks of aging.
SIRT6
SIRT6 is a multifunctional protein that has garnered significant attention for its role in aging and longevity. It is known to possess several enzymatic activities that contribute to its ability to protect cells, tissues, and organs from the effects of aging. One of the key mechanisms by which SIRT6 exerts its anti-aging effects is by promoting DNA repair. Studies have shown that SIRT6 is involved in the repair of DNA damage [43, 44, 45], which is a crucial aspect of maintaining cellular function and preventing the onset of age-related diseases. In addition to DNA repair, SIRT6 plays a critical role in maintaining the normal structure of chromosomes [46, 47, 48], which is essential for the proper functioning of cells throughout an individual's lifespan. Furthermore, SIRT6 is involved in regulating energy metabolism [49, 50], which is a vital process for maintaining cellular homeostasis and function. By influencing metabolic pathways, SIRT6 helps to ensure that cells can efficiently manage their energy requirements, thereby supporting overall health and longevity. SIRT6 also plays a role in regulating the senescence-associated secretory phenotype (SASP), which is the process by which senescent cells secrete pro-inflammatory cytokines, growth factors, and proteases that can contribute to tissue dysfunction and aging [51, 52]. By modulating SASP, SIRT6 helps to mitigate the detrimental effects of cellular senescence on the aging process. In addition to its roles in DNA repair, chromosome stability, metabolism, and SASP regulation, SIRT6 has been shown to inhibit immunosenescence, a process in which the immune system becomes less effective as we age [53]. Immunosenescence is associated with a decline in immune function, which can make the body more susceptible to infections, cancers, and other age-related diseases. By inhibiting immunosenescence, SIRT6 helps to maintain a robust immune system throughout the aging process. Moreover, SIRT6 has the ability to influence the differentiation and function of immune cells. It regulates post-translational modifications (PTMs) that affect various aspects of immune cell biology, including their differentiation, maturation, and function. SIRT6 also plays a role in immunometabolism, the process by which immune cells regulate their metabolic activity to support immune responses [54]. This makes SIRT6 an essential player in the immune system’s ability to adapt to different challenges throughout life. Despite these well-documented functions, further studies are required to fully understand the role of SIRT6 in regulating inflammation, particularly its impact on different immune cells in various diseases or at different stages of aging. Research is also needed to elucidate how SIRT6 influences the differentiation, maturation, and function of immune cells under different conditions. This will provide a deeper understanding of how SIRT6 may be leveraged to enhance immune function and improve health outcomes across the lifespan [54].
In addition to mitochondrial dysfunction and genomic instability, aging is also characterized by the damaging effects of oxidative stress and lipid peroxidation. 4-Hydroxy-2-nonenal (4-HNE), a byproduct of these processes, stands out as a significant biomarker linking oxidative damage, inflammation, and cellular aging.
4-Hydroxy-2-nonenal (4-HNE)
Building on the role of SIRT6 in preserving mitochondrial function and genomic stability, 4-Hydroxy-2-nonenal (4-HNE) emerges as a significant marker linking lipid peroxidation, oxidative stress, inflammation, and cellular aging. A major a, β-unsaturated aldehyde produced during lipid peroxidation, 4-HNE acts as a potent messenger in various signalling pathways [55]. Its binding to proteins (4-HNE-protein adducts) serves as a critical marker of lipid peroxidation, with levels increasing in brain tissues and fluids during aging. This accumulation is associated with hallmark aging disorders, particularly neurodegenerative diseases. Elevated 4-HNE inhibits telomerase and proteasomes, promoting telomere shortening and protein accumulation—key contributors to neurodegenerative and degenerative conditions.
A study by Maciejczyk et al. analyzing salivary and plasma biomarkers across 180 healthy individuals in six age groups (6–13, 14–19, 20–39, 40–59, 60–79, and 80–100 years) found that 4-HNE levels rise with age, influencing the expression of senescence-related pathways [56]. As a byproduct of lipid peroxidation caused by dyshomeostasis in reactive oxygen species (ROS), excessive 4-HNE exacerbates aging-related damage [55,57]. Elevated 4-HNE levels heighten the risk of inflammaging (chronic inflammation associated with aging) and several age-related disorders, including Alzheimer’s and Parkinson’s diseases, dry eye, macular degeneration, hearing loss, and cancer. These conditions arise due to the acceleration of oxidative stress-induced damage [57]. Factors contributing to abnormal 4-HNE levels include increased cellular oxidative stress, which disrupts the balance seen in healthy aging individuals. To mitigate these effects, interventions such as glutathione supplementation, an antioxidantrich diet, and regular physical activity are recommended to restore 4-HNE levels to a healthy range.
Building on the role of 4-HNE in linking oxidative stress, inflammation, and cellular aging, Taurine emerges as another key biomarker, addressing not only systemic inflammation but also cellular senescence, mitochondrial dysfunction, telomerase deficiency, and DNA damage.
Taurine
Taurine (2-aminoethanesulfonic acid) is a versatile biomarker that plays a pivotal role in multiple biological processes and aging mechanisms, such as cellular senescence, telomerase deficiency, mitochondrial dysfunction, DNA damage, and inflammation (Figure 2). Although taurine is the most abundant free amino acid in animal tissues, it is not incorporated into proteins. Its best-known biochemical role is as a precursor of the bile acid taurocholic acid [58]. In mammalian cells, taurine is synthesized from cysteine through the enzyme cysteine sulfinic acid decarboxylase (CSAD) [59].
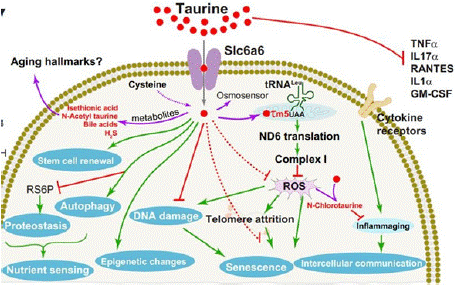
Figure 2: Singh et.al, illustrated effect of taurine and taurine derived
biomolecules (in red) on various hallmarks of aging.
With age, taurine levels naturally decline, exacerbating cellular aging processes and increasing susceptibility to conditions such as abdominal obesity, hypertension, inflammation, and type 2 diabetes. Studies have shown that restoring taurine levels can slow the aging process by mitigating its detrimental effects. The decline in taurine levels beyond normal rates observed in healthy aging individuals (where biological age aligns with chronological age) can result from deficiencies in key nutrients such as vitamin A, zinc, cysteine, or methionine. A reduced capacity to synthesize taurine from available precursors is another major factor contributing to its depletion with age [59]. To address this decline, taurine supplementation or increased intake of taurine-rich foods such as seafood, meats, legumes, and mollusks is recommended. Regular exercise can also help boost taurine levels. However, it is essential to note that individuals taking medications for low blood pressure should avoid taurine supplementation. Building on Taurine’s role in addressing inflammation, senescence, and DNA damage, mTOR emerges as a key regulator of aging, integrating growth and metabolic signals. While essential for repair and growth, chronic mTOR activation accelerates aging and inflammation, highlighting its critical role in balancing cellular homeostasis.
Mammalian/mechanistic Target of Rapamycin (mTOR)
Mammalian target of rapamycin (mTOR) is an evolutionarily conserved nutrient-sensing protein kinase that regulates growth, metabolism, and aging-related processes in eukaryotic cells [60]. mTOR is a central regulator that integrates nutrient availability with cellular processes such as cell growth, proliferation, and proteostasis. It plays a pivotal role in aging mechanisms, including nutrient sensing, maintenance of proteostasis, autophagy, mitochondrial function, cellular senescence, and stem cell decline [61].
Proteostasis, a critical aspect of cellular health, is regulated by mTOR through its influence on protein synthesis, degradation via proteasomal pathways or autophagy, and quality control mechanisms like the unfolded protein response (UPR) [61]. Dysregulation of mTOR signaling is associated with accelerated aging and age-related diseases due to its impact on these cellular pathways.
To mitigate the detrimental effects of chronic mTOR activation, approaches such as caloric restriction, pharmacological inhibitors like rapamycin, and strategies to enhance autophagy are being explored. These interventions aim to restore balance in mTOR signaling, thus promoting cellular homeostasis and potentially extending healthspan.
While mTOR highlights the complex interplay between cellular signaling and aging processes, the broader landscape of aging research presents significant challenges. Addressing these hurdles, such as identifying reliable biomarkers and understanding their multifaceted roles, will be crucial for uncovering new insights and shaping future directions in the field.
Current Challenges in the Field
Aging research faces significant hurdles that limit the applicability and generalizability of findings. Many existing studies suffer from small sample sizes and a lack of demographic diversity, reducing their ability to capture variations across populations. Furthermore, cross-sectional designs dominate the field, offering only snapshots of biological states rather than the dynamic insights gained from longitudinal studies.
Data integration presents another challenge; standardizing protocols and outputs across omics platforms remains difficult, complicating the comparison and consolidation of results. While genetics has been a predominant focus in understanding the aging process, the complexities of aging demand a more comprehensive approach. Developing a robust database that integrates multi-omics and real-time data is essential to provide a holistic view of the aging process. This approach will offer insights into the interplay of diverse factors shaping aging trajectories. Additionally, the absence of universally agreed-upon biomarker panels creates inconsistencies in defining reliable indicators for aging and healthspan.
Future Directions
The future of aging research lies in fostering multi-disciplinary collaborations to enable large-scale, longitudinal studies that account for diverse populations. Establishing standardized frameworks for data collection and analysis will be vital for harmonizing results across studies and omics platforms.
Researchers should prioritize under-researched areas, such as the interplay between biomarkers and lifestyle factors, to understand how interventions influence aging trajectories. Evaluating the long-term effects of these interventions on biomarker profiles will yield actionable insights for developing effective anti-aging therapies and healthspan optimization strategies. Moreover, integrating genomic, proteomic, metabolomic, and functional health biomarkers into comprehensive panels will enhance our understanding of the aging process.
Conclusion
This review highlights the critical role of biomarkers in unravelling the complexities of the aging process, focusing on a select few genomic and proteomic biomarkers. While these biomarkers provide valuable insights into aging mechanisms, a more extensive panel of genomic, proteomic, metabolomic, and functional health biomarkers is needed to gain a comprehensive understanding. Future reviews will explore these broader panels, emphasizing the need to correlate these biomarkers with the numerous factors influencing aging.
Bringing all this information together will allow researchers to evaluate these correlations and uncover more profound insights into the aging process. Such an integrated approach will not only advance our understanding of aging but also provide leverage to address its hallmarks effectively. A concerted effort toward collaborative, large- scale, and interdisciplinary research will pave the way for breakthroughs in precision strategies to extend healthspan and improve quality of life.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. Suresh Poosala envisioned this approach of understanding the interplay of genomic, proteomic and functional biomarkers having a key mechanistic role in the aging of an individual and identified several biomarkers with such key interplay influencing characteristics. Nikita Naik assimilated and wrote the manuscript.
References
- Salignon J, Faridani OR, Miliotis T, Janssens GE, Chen P, Zarrouki B, et al. Age prediction from human blood plasma using proteomic and small RNA data: a comparative analysis. Aging (Albany NY). 2023; 15: 5240-5265.
- Vitetta L & Anton B. Lifestyle and nutrition, caloric restriction, mitochondrial health and hormones: scientific interventions for anti-aging. Clinical Interventions in Aging. 2007; 2: 537–543.
- Bafei SEC, Shen C. Biomarkers selection and mathematical modeling in biological age estimation. npj Aging. 2023; 9: 13.
- Jylhävä J, Pedersen NL & Hägg S. Biological Age Predictors. EBioMedicine. 2017; 21: 29–36.
- Diebel LWM & Rockwood K. Determination of Biological Age: Geriatric Assessment vs Biological Biomarkers. Current Oncology Reports. 2021; 23.
- Jaiswal, Siddhartha, and Benjamin L. Ebert. “Clonal hematopoiesis in human aging and disease.” Science. 2019; 366.6465: eaan4673.
- Moldogazieva, Nurbubu T, et al. “Oxidative stress and advanced lipoxidation and glycation end products (ALEs and AGEs) in aging and age-related diseases.” Oxidative medicine and cellular longevity. 2019.
- Boss GR and JE Seegmiller. “Age-related physiological changes and their clinical significance.” The Western journal of medicine. 1981; 135: 434-440.
- Warner, Huber R, et al. “Program for testing biological interventions to promote healthy aging.” Mechanisms of Ageing and Development. 2000; 115: 199-207.
- Yu Mengdi, et al. “Key signaling pathways in aging and potential interventions for healthy aging.” Cells. 2021; 10: 660.
- López-Otín C, Blasco MA, Partridge L, Serrano M & Kroemer G. Hallmarks of aging: An expanding universe. In Cell. 2023; 186: 243–278.
- Lo´pez-Oti´n C, Blasco MA, Partridge L, Serrano M and Kroemer G. The hallmarks of aging. Cell. 2013; 153: 1194–1217.
- Baker GT. 3rd & Sprott RL. Biomarkers of aging. Exp. Gerontol. 1988; 23: 223–239.
- Herskind AM, McGue M, Holm NV, Sørensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet. 1996; 97: 319-323.
- vB Hjelmborg J, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006; 119: 312.
- Deelen J. et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum. Mol. Genet. 2014; 23: 4420–4432.
- Quiles J, Cabrera M, Jones J, Tsapekos M, Caturla N. In Vitro Determination of the Skin Anti-Aging Potential of Four Component Plant-Based Ingredient. Molecules. 2022; 27: 8101.
- Demanelis K, Jasmine F, Chen LS, Chernoff M, Tong L, Delgado D, et al. Determinants of telomere length across human tissues. Science. 2020; 369: eaaz6876.
- von Zglinicki T, Wan T, Miwa S. Senescence in Post-Mitotic Cells: A Driver of Aging? Antioxid Redox Signal. 2021; 34: 308-323.
- Der G, Batty GD, Benzeval M, Deary IJ, Green MJ, McGlynn L, et al. Is telomere length a biomarker for aging: cross-sectional evidence from the west of Scotland? PLoS ONE. 2012; 7: e45166.
- Fragkiadaki P, Nikitovic D, Kalliantasi K, Sarandi E, Thanasoula M, Stivaktakis PD, et al. Telomere length and telomerase activity in osteoporosis and osteoarthritis. Exp Ther Med. 2020; 19: 1626-1632.
- Vaiserman A, Krasnienkov D. Telomere Length as a Marker of Biological Age: State-of-the-Art, Open Issues, and Future Perspectives. Front Genet. 2021; 11: 630186.
- Habibi N, Bianco-Miotto T, Phoi YY, Jankovic-Karasoulos T, Roberts CT and Grieger JA. Maternal diet and offspring telomere length: a systematic review. Nutr. Rev. 2021.
- Gorenjak V, Petrelis AM, Stathopoulou MG and Visvikis-Siest S. Telomere length determinants in childhood. Clin. Chem. Lab. Med. 2020; 58: 162–177.
- Beijers R, Hartman S, Shalev I, Hastings W, Mattern BC, de Weerth C, et al. Testing three hypotheses about effects of sensitive-insensitive parenting on telomeres. Dev Psychol. 2020; 56: 237–250.
- Ilmonen P, Kotrschal A and Penn DJ. Telomere attrition due to infection. PLoS ONE. 2008; 3: e2143.
- Boccardi M and Boccardi V. Psychological wellbeing and healthy aging: focus on telomeres. Geriatrics. 2019; 4: 25.
- Mayer SE, Prather AA, Puterman E, Lin J, Arenander J, Coccia M, et al. Cumulative lifetime stress exposure and leukocyte telomere length attrition: the unique role of stressor duration and exposure timing. Psychoneuroendocrinology. 2019; 104: 210–218.
- Canudas S, Becerra-Tomás N, Hernández-Alonso P, Galié S, Leung C, Crous-Bou M, et al. Mediterranean diet and telomere length: a systematic review and meta-analysis. Adv Nutr. 2020; 11: 1544–1554.
- Semeraro MD, Smith C, Kaiser M, Levinger I, Duque G, Gruber HJ, et al. Physical activity, a modulator of aging through effects on telomere biology. Aging. 2020; 12: 13803–13823.
- Astuti Y, Wardhana A, Watkins J, Wulaningsih W and PILAR Research Network. Cigarette smoking and telomere length: a systematic review of 84 studies and meta-analysis. Environ. Res. 2017; 158: 480–489.
- Dixit S, Whooley MA, Vittinghoff E, Roberts JD, Heckbert SR, Fitzpatrick AL, et al. Alcohol consumption and leukocyte telomere length. Sci Rep. 2019; 9: 1404.
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genomewide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013; 49: 359–367.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14: R115.
- Jylhävä J, Pedersen NL & Hägg S. Biological Age Predictors. In EBioMedicine. 2017; 21: 29–36.
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-anal ysis predicting time to death. Aging (Albany NY). 2016; 8: 1844–1865.
- Anderson NL, Anderson NG. Proteome and proteomics: new tech nologies, new concepts, and new words. Electrophoresis. 1998; 19: 1853–1861.
- Ignjatovic V, Lai C, Summerhayes R, Mathesius U, Tawfilis S, Perugini MA, et al. Age-related differences in plasma proteins: how plasma proteins change from neonates to adults. PLoS One. 2011; 6: e17213.
- Menni C, Kiddle SJ, Mangino M, Viñuela A, Psatha M, Steves C, et al. Circulating proteomic signatures of chronological age. J Gerontol A Biol Sci Med Sci. 2015; 70: 809–816.
- Ulgherait M, Rana A, Rera M, Graniel J, Walker DW. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 2014; 8: 1767-1780.
- Fenouille N, Nascimbeni AC, Botti-Millet J, Dupont N, Morel E, Codogno P. To be or not to be cell autonomous? Autophagy says both. Essays Biochem. 2017; 61: 649-661.
- Lim CT, Lolli F, Thomas JD, Kola B & Korbonits M. Chapter Seventeen - Measurement of AMP-Activated Protein Kinase Activity and Expression in Response to Ghrelin. In M. Kojima & K. B. T.-M. in E. Kangawa (Eds.), Ghrelin. 2012; 514: 271–287.
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic Instability and Aging-Like Phenotype in the Absence of Mammalian Sirt6. Cell. 2006; 124: 315–329.
- Xu Z, Zhang L, Zhang W, Meng D, Zhang H, Jiang Y, et al. SIRT6Rescues the Age Related Decline in Base Excision Repair in a PARP1-Dependent Manner. Cell Cycle. 2015; 14: 269–276.
- Geng A, Tang H, Huang J, Qian Z, Qin N, Yao Y, et al. The Deacetylase SIRT6 Promotes the Repair of UV-Induced DNA Damage by Targeting Ddb2. Nucleic Acids Res. 2020; 48: 9181–9194.
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappaB-Dependent Gene Expression and Organismal Life Span. Cell. 2009; 136: 62–74.
- Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is Required for Maintenance of Telomere Position Effect in Human Cells. Nat Commun. 2011; 2: 433.
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. SIRT6 Represses LINE1 Retrotransposons by Ribosylating KAP1 But This Repression Fails With Stress and Age. Nat Commun. 2014; 5: 5011.
- Roichman A, Elhanati S, Aon MA, Abramovich I, Di Francesco A, Shahar Y, et al. Restoration of Energy Homeostasis by SIRT6 Extends Healthy Lifespan. Nat Commun. 2021; 12: 3208.
- Wiley CD, Campisi J. The Metabolic Roots of Senescence: Mechanisms and Opportunities for Intervention. Nat Metab. 2021; 3: 1290–1301.
- Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. Cellular Senescence and the Senescent Secretory Phenotype: Therapeutic Opportunities. J Clin Invest. 2013; 123: 966–972.
- Van Gool F, Galli M, Gueydan C, Kruys V, Prevot PP, Bedalov A, et al. Intracellular NAD Levels Regulate Tumor Necrosis Factor Protein Synthesis in a Sirtuin-Dependent Manner. Nat Med. 2009; 15: 206–210.
- Lasiglie D, Boero S, Bauer I, Morando S, Damonte P, Cea M, et al. Sirt6 Regulates Dendritic Cell Differentiation, Maturation, and Function. Aging (Albany NY). 2016; 8: 34–49.
- Li Y, Jin J & Wang Y. SIRT6 Widely Regulates Aging, Immunity, and Cancer. In Frontiers in Oncology. 2022; 12.
- Zhang H, Forman HJ. 4-hydroxynonenal-mediated signaling and aging. Free Radic Biol Med. 2017; 111: 219-225.
- Maciejczyk M, Nesterowicz M, Szulimowska J, Zalewska A. Oxidation, Glycation, and Carbamylation of Salivary Biomolecules in Healthy Children, Adults, and the Elderly: Can Saliva Be Used in the Assessment of Aging? J Inflamm Res. 2022; 15: 2051-2073.
- Li Y, Zhao T, Li J, Xia M, Li Y, Wang X, et al. Oxidative Stress and 4-hydroxy- 2-nonenal (4-HNE): Implications in the Pathogenesis and Treatment of Agingrelated Diseases. J Immunol Res. 2022; 2022: 2233906.
- Massie HR, Williams TR & DeWolfe LK. Changes in taurine in aging fruit flies and mice. Experimental Gerontology. 1989; 24: 57–65.
- Singh P, Gollapalli K, Mangiola S, Schranner D, Yusuf MA, Chamoli M, et al. Taurine deficiency as a driver of aging. Science. 2023; 380.
- Stallone G, Infante B, Prisciandaro C & Grandaliano G. MTOR and aging: An old-fashioned dress. In International Journal of Molecular Sciences. 2019; 20.
- Papadopoli D, Boulay K, Kazak L, Pollak M, Mallette FA, Topisirovic I & Hulea L. Mtor as a central regulator of lifespan and aging. F1000Research. 2019; 8.