
Research Article
Austin J Clin Cardiolog. 2023; 9(2): 1107.
Genetic Lineage Traces the Differentiation Fate of Epicardial Cells during Heart Development
FR Lu*; XJ Yang
College of Life Science and Technolog, Jinan University, Guangzhou, 510632, Guangdong, China
*Corresponding author: FR Lu College of Life Science and Technolog, Jinan University, Guangzhou, 510632, Guangdong, China. Tel: +8618337319163 Email: [email protected]
Received: November 06, 2023 Accepted: December 11, 2023 Published: December 18, 2023
Abstract
Objective: The genetic lineage tracing method was used to examine the Epithelial-Mesenchymal Transition (EMT) process and the contribution of epicardial cells to mesenchymal cells at various stages of fetal heart development.
Methods: In Wt1-CreER; R26-tdTomato transgenic mice, tamoxifen was utilized to promote the tagging of epicardial cells with tdTomato fluorescence at E10. At E11.5, E12.5, and E16.5, embryonic hearts were harvested and photographed using confocal fluorescence microscopy and stereomicroscopy.
Results: According to the findings, the tdTomato+ cells at E11.5 were still in the epicardium and had not yet moved into the myocardium. Epicardial cells began to separate from the epicardium and give rise to epicardial-derived cells at embryonic day 12.5 (E12.5). On the valve primordium, fibroblasts generated from epicardium have been found. By E16.5, many epicardial cells had moved into the myocardium and formed fibroblasts, mesenchymal cells, vascular smooth muscle cells, as well as migrated into the ventricular septum and valves, contributing to their growth and creation.
Conclusions: The contribution of epicardial cells to mesenchymal cells during development is shown by genetic lineage tracing, opening up possibilities and offering references for creating relevant treatment approaches based on epicardial cells.
Keywords: Lineage tracing; Epithelial-to-mesenchymal transition; Valve; Heart
Introduction
Heart diseases, including heart failure, myocardial infarction, and coronary artery disease, have become significant global health issues [1-2]. Following heart damage, activated epicardium-derived cells (EPDCs) can invade the injury site, promoting the proliferation of coronary artery endothelial cells, regenerating myocardial cells, and forming fibroblasts to maintain normal heart function [3]. Embryonic epicardial cells possess greater differentiation potential compared to adult epicardial cells, and transplantation of epicardial cells derived from embryonic stem cells has emerged as a promising therapy for various heart diseases. Therefore, the use of lineage tracing systems to track the fate of embryonic epicardial cells holds significant potential for the treatment of heart disease [4].
The Cre-loxP system is a widely used, powerful technology for mammalian gene editing. Concerning the mechanism of the Cre-loxP system, a single Cre recombinase recognizes two repeated loxP site, then the Cre excises the loxP-flanked DNA [5]. To achieve more accurate genetic functional studies and clinical applications using the Cre-loxP system,a more sophisticated technique was required that controled the Cre activation at a precise time and in an specific cell. An inducible Cre system is controlled by cell-specific regulatory elements (promoters and enhancers) and inducible way by exogenous inducers such as tam [6].
EMT refers to epithelial cells losing their epithelial features and acquiring mesenchymal characteristics with migratory potential. EMT has been shown to be essential for both developmental and pathological processes, including embryo morphogenesis, wound healing, tissue fibrosis, and cancer. Heart formation involves a series of epithelial-mesenchymal transition and reverse EMT processes, with almost all cardiac and non-cardiac cells generated through EMT. During heart development, EPDCS can migrate into the myocardium and differentiate into various cell types, including smooth muscle cells that comprise the coronary artery, acute fibroblasts, and so on [7]. EPDCs also contribute to the development of the atrioventricular valves and ventricular septum, with fibroblasts derived from the epicardium preferentially participating in the formation and development of the wall leaflets of the atrioventricular valve [8-9].
The epicardium is typically distinguished by the expression of transcription factors T-box18 (Tbx18),Wilm tumor gene1(Wt1), and Transcription factor 21(Tcf21). While Tbx18 is known to be involved in heart development, it is not essential for the formation of cardiac chambers and coronary vessels and may also be expressed in the myocardium, potentially confounding experimental results [10]. Tcf21, on the other hand, plays an important role in cell differentiation and growth processes and is expressed in cardiac mesenchymal cells, making it a commonly used marker in zebrafish studies [11]. In this study, we focused on characterizing the fate of epicardial-derived cells during the epithelial-to-mesenchymal transition in embryonic heart development, using Wt1 to drive the Cre-LoxP system. Wt1 promotes epicardial EMT through Wnt and retinoic acid signaling pathways, and its inactivation can lead to the abnormal formation of EPDCs and affect normal heart development. Our findings provide valuable insights into the role of epicardial cells in the development of valvular and ventricular septa.
Materials and Methods
Animal Studies and Ethics Statement
All Animals were used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. The study was approved by the Institutional Animal Care and Use Committee of Jinan University (approval number: 20220613-16). The Wt1-CreER;R26-tdTomato knock-in mouse line was generated by homologous recombination using CRISPR/Cas9 methods. Wt1-CreER; Rosa26-tdTomato mice lines were maintained on a C129/C57BL6/J-mixed background.
We administered 5mg tam (Sigma) by gavage to pregnant mice at E10.5 to induce Cre.
Tissue Collection and Immunostaining
Mouse embryonic hearts were collected in PBS and then washed gently to remove blood. They were then fixed in 4% PFA for 15-60 min at 4°C depending on tissue size. After washing several times in PBS, we used fluorescence microscopy (Leica M205 FA/DFC 7000T) to take photographs of embryonic heart with indicated fluorescence reporters.After that, the embryonic hearts were dehydrated in 30% sucrose/PBS for several hours or overnight at 4°C, and then embedded in OCT (Sakura) the next day. Frozen sections of 6-10 μm were collected on slides. For immunostaining, tissue sections were first washed in PBS to remove OCT, then blocked in DAPI for 30 min at room temperature and then incubated with primary antibodies overnight at 4°C. Primary antibodies against the following proteins and dilution were used:Wt1 (Abcam,ab89901;1:200), a-smooth muscle actin (aSMA) (Abcam, ab5694; 1:500), platelet-derived growth factor receptor β (PDGFRβ) (eBioscience, 14-1402-82; 1:2000), platelet-derived growth factor receptor a (PDGFRa) (R&D, AF062; 1:1000), RFP Antibody pre-adsorbed(Rockland, 600-401-379; 1:500) and ImmPRESS Horse Anti-Rabbit IgG (Vector lab, MP-7401; 1:1). The next day, sections were washed with PBS several times and then incubated with Alexa fluorescence secondary antibodiesat room temperature for 30 min in dark.After washing several times in PBS, all slides were mounted with mounting medium. For weak signals, HRP-conjugated secondary antibodies were used to amplify the signals. Images were acquired using an Olympus confocal microscopy system (FV3000).
Genomic PCR
Genomic DNA was extracted from mouse tail,Tissues were lysed in lysis buffer (100 μg/ml proteinase K) overnight at 55°C and the mixture was centrifuged at the maximum speed of 20,000 g for 8 min to obtain supernatant with genomic DNA the next day. DNA was precipitated using isopropanol, and then washed in 70% ethanol by centrifugation at 20,000g for 3 min.
All the mice or embryos were genotyped using genomic PCR to distinguish knock-in allele from wild-type allele.For the R26-tdToamto line, primers 5'-TCCCGACAAAACCGAAAATCTGTGG-3' and 5'-TGCATCGCATTGTCTGAGTAGG-3' were used to detect the R26-tdToamt positive allele, and 5'-TCCCGACAAAACCGAAAATCTGTGG-3',5'-GGGGCGTGCTGAGCCAGACCTCCAT-3' were
used to detect the wild-type allele.For the Wt1-Cre line, primers 5'-GGCTTAAAGGCTAACCTGGTGTG-3' and 5'-GGAGCGGGAGAAATGGATATG-3' were used to
detect the Wt1 positive allele, and 5'-CCAAGTCCAGCGCCGAGAAT-3' and 5'-TGTCCATCAGGTTCTTGCGA-3' were used to detect the wild-type allele.
Statistical Analysis
All data were obtained from three to more independent experiments, as indicated in each figure legend and were presented as mean±Standard Deviation .Two-tailed Student’ s t tests were used to calculate statistical significance with p values. P value<0.05 was considered statistically significant, ns is non-statistically significant. Graphs were generated using GraphPad Prism 9.0.0 software.
Results
The Cre-LoxP System Functions After Tamoxifen
The Cre-LoxP system is commonly used for tracing mammalian cell lineages and has been widely employed in lineage tracing studies of various organs and cell types [12-13]. Notably, Cre recombinase is dependent on tamoxifen induction and only translocates into the nucleus to mediate recombination under tamoxifen-induced conditions (Figure 1A,1B).
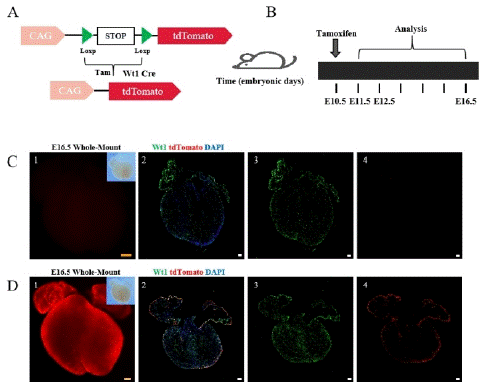
Figure 1: The working principle of the Cre-LoxP system and research strategy.
Note: (A) The working principle of the Cre-Loxp system. (B) Research strategy diagram. Tamoxifen (5mg) was administered to pregnant a mouse at E10.5, and Wt1-CreER; R26-tdTomato embryonic hearts were collected at E11.5, E12.5, and E16.5. (C) 1-4, Bright field and fluorescence views of E16.5 embryonic hearts without tamoxifen induction (no tam). Wt1 (green) is a specific marker for epicardial cells. tdTomato labeling epicardial cells and EPDCS, DAPI (blue) as a DNA fluorescent dye. (D) 1-4 Bright field and fluorescence views of E16.5 embryonic hearts with tamoxifen induction (tam). Scale bars: 250 μm (yellow) and 100 μm (white).
Comparison of embryonic hearts with and without tamoxifen induction revealed weak fluorescence and few tdTomato+ cells in the non-induced group (Figure 1C), while high expression of tdTomato was observed throughout the heart in the tamoxifen-induced group (Figure 1D).These results indicate that the Cre recombinase only becomes active upon tamoxifen induction.
Epicardial Cells did not Migrate at E11.5
The epicardium had formed on the surfaces of the embryonic heart at E11.5, the valves were endocardial cushion, and the interventricular septum was not fully developed at this phase. To track the expression and distribution of epicardial cells at this phase,after tamoxifen induction at E10.5, the Cre-loxp system was activated, and epicardial cells were labeled with tdTomato fluorescent protein.
It was observed that tdTomato+ and Wt1+ were coexpressed only in epicardial cells. The tdTomato+ cells did not invade the myocardium, indicating that the epicardial cells did not detach from the epicardial region and did not produce EPDCs at this time (Figure2). During this period, the precursor epicardium-to-epicardium transformation was occurring, which is the main source of epicardial cell formation[14].
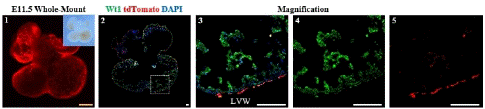
Figure 2: Spatial Distribution of Epicardial Cells in the E11.5 Embryonic Heart.
Note: 1. Whole-mount Bright field and fluorescence views of E11.5 embryonic hearts 2. Immunofluorescence view of heart sections. 3-5: Higher magnification immunofluorescence views of heart sections. Left Ventricular Wall (LVW). Scale bars: 250μm (yellow) and 100μm (white).
The Epicardial Cells Begin to Migrate and Form Fibroblast at E12.5
At this phase, the endocardial cushions gradually fuse, and the atrioventricular canal divides into the left and right ventricles, forming the atrioventricular valve. By tracking the expression distribution of epicardial cells at E12.5, it was found that tdTomato+ cells began to detach from the epicardium and invade the myocardium,indicating the beginning of the EMT process.
Using confocal microscopy and fluorescent stereomicroscopy, it was observed that tdTomato+ cells had begun to express in areas other than the epicardium at this phase, with the earliest detection found in the region of the valve primordia, and more expression in the wall leaflet of the tricuspid valve primordia.
It is worth mentioning that some of the tdTomato+ cells at the site of the valve primordia merged with PDGFRa+ cells, indicating that epicardial cells began to undergo EMT and form fibroblasts (Figure 3).
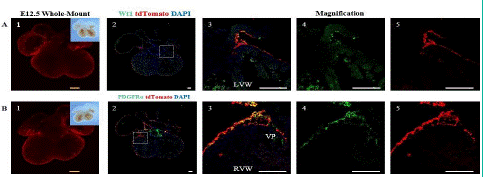
Figure 3: Spatial Distribution of Epicardial Cells and Fibroblasts in the E12.5 Embryonic Heart.
Note: (A) 1.Whole-mount bright field and fluorescence views of the E12.5 ventral embryonic heart. (A) 2. immunofluorescence views of heart sections. (A) 3-5: Higher magnification immunofluorescence views of heart sections. (B) 1. Whole-mount bright field and fluorescence images of the E12.5 dorsal embryonic heart. (B) 2.immunofluorescence views of heart sections. PDGFRa (green) is a specific marker for fibroblasts.td Tomato (red), and DAPI (blue). (B) 3-5: Higher magnification immunofluorescence views of heart sections. The Right Ventricular Wall (RVW), Valve Primordial (VP). Scale bars: 250μm (yellow) and 100μm (white).
At E16.5, Epicardial Cells Differentiate into Fibroblasts, Vascular Smooth Muscle Cells, and Mesenchymal Cells
To track the expression distribution and differentiation of epicardial cells at E16.5, we added two new immunofluorescence markers in addition to epicardial and fibroblast markers: aSMA and PDGFRβ [15-17].
Through immunofluorescence staining, we observed a large number of tdTomato+ cells migrating into the myocardium and dividing into fibroblasts, vascular smooth muscle cells, and mesenchymal cells. Some undifferentiated EPDCs will remain in the epicardial space and myocardial tissue and retain the expression characteristics of Wt1 (Figure 4A). Large circular vascular signals that merged with tdTomato+ cells appeared as yellow signals (Figure 4B). These larger vessels are usually detected in the right ventricle. We also found that tdTomato+ cells merged with signals from fibroblasts and mesenchymal cells (Figure 4C, 4D), consistent with previous studies [18-19]. By statistical analysis, approximately 7.47±1.48%, 33.47±4.19%, and 42.77±2.15% of EPDCs differentiated into vascular smooth muscle cells, fibroblasts, and mesenchymal cells (Figure 5).
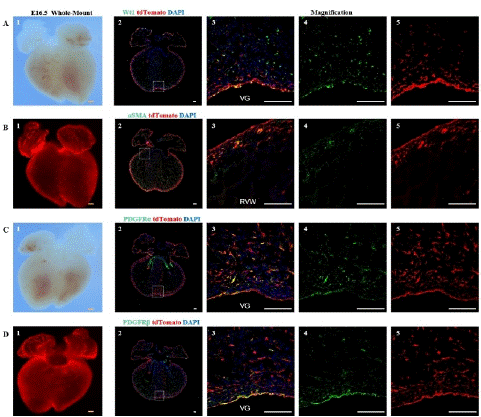
Figure 4: Spatial distribution of epicardial cells, fibroblasts, vascular smooth muscle cells, and mesenchymal cells at E16.5
Note: (A) 1, Bright-field image of the ventral view of an E16.5 embryo heart. (A) 2, Immunofluorescence image of E16.5 heart sections,. Wt1 (green), tdTomato (red), DAPI (blue); (A) 3-5, Higher magnification immunofluorescence images of E16.5 heart sections, VG (Ventricular Groove). (B) 1, Fluorescence image of the ventral view of an E16.5 embryo heart. (B) 2, immunofluorescence image E16.5 heart sections. aSMA (green), tdTomato (red), DAPI (blue); (B) 3-5, Higher magnification immunofluorescence images of E16.5 heart sections, RVW (Right Ventricular Wall). (C) 1, Bright-field image of the dorsal view of an E16.5 embryo heart. (C) 2, immunofluorescence image of E16.5 heart sections. PDGFRa (green), tdTomato (red), DAPI (blue); (C) 3-5, Higher magnification immunofluorescence images of E16.5 heart sections. (D) 1, Fluorescence image of the dorsal view of an E16.5 embryo heart. (D) 2, immunofluorescence image of E16.5 heart sections. PDGFRβ (green) is a marker for mesenchymal cells, tdTomato (red), DAPI (blue); (D) 3-5, Higher magnification immunofluorescence images of E16.5 heart sections,. Scale bars: 250μm (yellow) and 100μm (white).
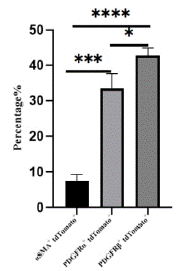
Figure 5: Statistical analysis of vascular smooth muscle cells, fibroblasts and mesenchymal cells.
Note: The proportion of aSMA+ tdTomato+, PDGFRa+tdTomato+ and PDGFRβ+ tdTomato+ cells in tdTomato+ cells were counted, sections n=3, totaling about 3000 cells.
*P<0.05,***P<0.001,****P<0.0001.
Involvement of Epicardial Cells in the Development and Formation of Valve and the Ventricular Septum
In many previous studies, it has been shown that epicardial cells play an important role in valve mesenchymal cell development. At E12.5, endocardium-derived cells constituted almost all of the valve mesenchyme in the leaflets of the mitral valve, while EDPCs contributed less [20].
At E14.5, EPDCs began to migrate into the proximal leaflets, and more EPDCs could be detected at E17.5. EPDCs can also differentiate into mesenchymal cells through signaling pathways and participate in the development and formation of the ventricular septum [21]. The expression of these markers in the valves and ventricular septum was analyzed by co-localization with tdTomato+ cells.
At E16.5, this study found that epicardial cells could migrate to the valves and ventricular septum (Figure 6). The different composition of the two leaflets of the tricuspid and mitral valves may be due to their different anatomical positions during heart development.
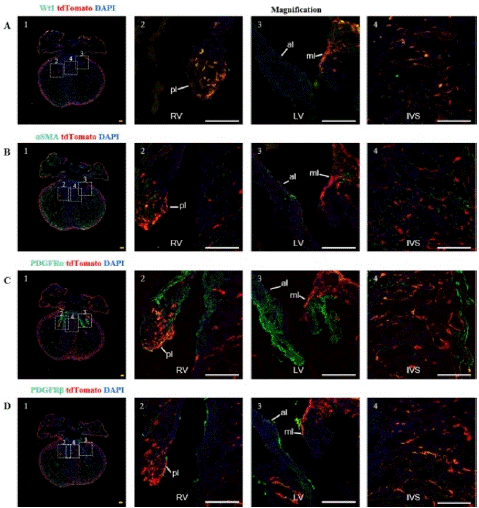
Figure 6: Spatial distribution of epicardial cells and EPDCs in the valves and ventricular septum at E16.5.
Note: (A) 1, immunofluorescence image of E16.5 heart sections, (A) 2, Higher magnification immunofluorescence views of tricuspid valve. (A) 3, Higher magnification immunofluorescence views of mitral valve; (A) 4, Higher magnification immunofluorescence views of interventricular septum; RV (Right Ventricle), LV (Left Ventricle), IVS (Interventricular Septum), pl (Parietal Leaflet, tricuspid valve), ml (Mitral Leaflet, mitral valve), al (Aortic Leaflet). (B)-(D)1-4 Same as group A except for maker. Scale bar: 100μm.
The parietal leaflet of the tricuspid valve attaches to the wall of the right ventricle, while the parietal leaflet of the mitral valve attaches to the wall of the left ventricle, where EPDCs mainly distribute. Approximately 32.57±0.5% of cells in the TV were derived from the epicardium, but most of these epicardial cells did not undergo conversion in the tricuspid valve and still retained epicardial cell characteristics. Only about 7.86±1.45% of cells were epicardium-derived mesenchymal cells in TV, including 2.53±1.79% fibroblasts. There were slightly more epicardial-derived fibroblasts in the MV than TV, accounting for about 5.23±0.07%. About 7.6±2.42% of cells were epicardium-derived mesenchymal cells in the MV. At E16.5, epicardial cells participated in the development and formation of valve cells, but at this time, most of the mesenchymal cells in the valves came from endocardial cells, and the role of epicardial cells was relatively small. In the ventricular septum, about 10% to 20% of cells were derived from the epicardium, among which about 9.23±0.64% formed mesenchymal cells, with fibroblasts accounting for the vast majority and the remaining cells retaining epicardial cell characteristics (Figure 7).
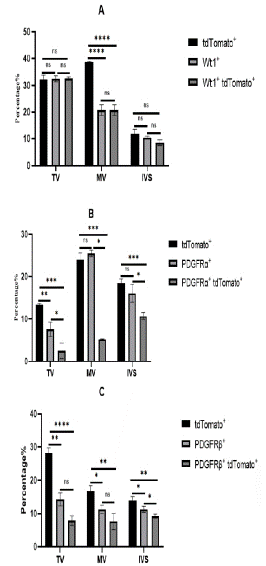
Figure 7: Statistical analysis of EPDCS in TV, MV, IVS.
Note: (A) Analyze the expression distribution of epicardial cells in TV, MV and IVS. Tricuspid Valve (TV), MV (Mitral Valve) and IVS (Interventricular Septum) interventricular septum (n=3). The total number is about 10,000 cells.* P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (B) The expression distribution of fibroblasts in TV, MV and IVS was statistically analyzed. (C) The expression distribution of mesenchymal cells in TV, MV, IVS was statistically analyzed.
Discussion
In this study, we generated a Wt1-CreER; R26-tdTomato mouse and tracked the dynamic changes in epicardial cell formation into various mesenchymal cells at different phase. However, some researchers have pointed out that the lack of exogenous tamoxifen may still activate Cre recombinase activity spontaneously due to endogenous estrogen or other factors, which may affect the research results [22]. We compared embryonic hearts induced with and without tamoxifen and confirmed the reliability of the Cre recombinase, showing that endogenous estrogen or other factors did not affect the experimental results.
At E10 to E10.5, the precursor epicardium gradually disappears, and the epicardium gradually forms, so tamoxifen was administered to a pregnant mouse at E10.5. By around E11.5, the precursor epicardium had disappeared, and the epicardium had formed and covered the entire outer layer of the heart. At E12.5, epicardial cells began to invade the myocardium. E16.5 is the early stage of coronary artery formation in the developing heart.
Before E12.5, the epicardial precursor cells mainly underwent transformation into epicardial cells in the epicardial region of the embryonic heart, which was the main source of epicardial cells [23]. By E12.5, some epicardial cells began to detach from their original positions and migrate into the myocardium, and they were first detected in the valvular primordia. At E16.5, a large number of epicardial cells migrated into the myocardium, forming mesenchymal cells.
However, some EPDCs that migrated to the ventricular wall retained expression of Wt1. The persistence of Wt1 expression may be related to the maintenance of the undifferentiated stem cell state in these EPDCs and the reactivation of EMT. During this period, epicardial cells also played an important role in the development and formation of valves and the ventricular septum [24]. Many EPDCs in the valves remained undifferentiated into mesenchymal cells, and at this time, mesenchymal cells in the valves may have been mainly derived from endocardial cells. Most epicardially-derived cells in the ventricular septum formed fibroblasts. Compared with using stem cell transplantation to induce myocardial regeneration for treating damaged hearts, epicardial progenitor cells have the advantages of clear differentiation direction and high safety. Studies have shown that combined transplantation of embryonic stem cell-derived cardiomyocytes and epicardial cells can help organs recover from heart attack injuries, and epicardial cells can help transplanted cardiomyocytes survive longer [25]. Therefore, further research on the EMT process of embryonic and adult epicardial cells will be of great significance for drug development, developing new medical approaches and methods, and treating various congenital heart diseases, such as ventricular septal defect, and many acquired heart diseases [26].
The Cre-LoxP system, as one of the commonly used tools for tracing mammalian cell lineages, has been widely used in lineage tracing of various organs and cells, but it still has limitations.The EMT process of epicardial cells requires the activation of various transcription factors, such as Snail and E-cadherin, which are necessary for epicardial cells to transform into cardiovascular progenitor cells [27]. These transcription factors can be considered markers of the EMT process. With the advancement of technology, more precise dual recombinase lineage tracing techniques have emerged, which can make the epicardial EMT process more accurate and detailed in future studies. In summary, this study used Wt1 as a marker for epicardial cells and revealed the contribution of epicardial cells to mesenchymal cells at different phases through genetic lineage tracing of cell fate. The study also tracked the involvement of epicardial cells in valve and interventricular septum development and formation. The contribution of epicardial cells to mesenchymal cells during development can provide a reference for the selection of transplantation of differentiated epicardial cells in the treatment of heart disease.
Author Statements
Acknowledgments
Xiaojie Yang has made a significant scientific contribution to the research in the manuscript, and agreed to be Co-first author. We thank all lab members (Xueying Tian, Tingting Han) for helpful discussions and excellent technical suppor. We also thank Shanghai Biomodel Organism Co. Ltd. for mouse generation and the Jinan University start-up fund strong support for this research. I have used Youdao Translation software to edit or translate my manuscripts.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Funding Statement
The Jinan University start-up fund is free of conflict.
References
- Taylor CJ, Hobbs FD. Heart failure therapy in patients with coronary artery disease. Curr Opin Pharmacol. 2013; 13: 205-9.
- He L, Nguyen NB, Ardehali R, Zhou B. Heart regeneration by endogenous stem cells and cardiomyocyte proliferation: controversy, fallacy, and progress. Circulation. 2020; 142: 275-91.
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011; 121: 1894-904.
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006; 127: 607-19.
- Kim H, Kim M, Im SK, Fang S. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res. 2018; 34: 147-59.
- He L, Li Y, Li Y, Pu W, Huang X, Tian X, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med. 2017; 23: 1488-98.
- Kässmeyer S, Plendl J, Custodis P, Bahramsoltani M. New insights in vascular development: vasculogenesis and endothelial progenitor cells. Anat Histol Embryol. 2009; 38: 1-11.
- Webb S, Brown NA, Anderson RH. Formation of the atrioventricular septal structures in the normal mouse. Circ Res. 1998; 82: 645-56.
- Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006; 98: 1547-54.
- Foster DB, Gu JM, Kim EH, Wolfson DW, O’Meally R, Cole RN, et al. Tbx18 orchestrates cytostructural transdifferentiation of cardiomyocytes to pacemaker cells by recruiting the epithelial-mesenchymal transition program. J Proteome Res. 2022; 21: 2277-92.
- Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019; 25: 1280-9.
- Liu Q, Zhang H, Tian X, He L, Huang X, Tan Z, et al. Smooth muscle origin of postnatal 2nd CVP is pre-determined in early embryo. Biochem Biophys Res Commun. 2016; 471: 430-6.
- He L, Li Y, Huang X, Li Y, Pu W, Tian X, et al. Genetic lineage tracing of resident stem cells by DeaLT. Nat Protoc. 2018; 13: 2217-46.
- Sacks D, Baxter B, Campbell BCV, Carpenter JS, Cognard C, et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. 2018; 13: 612-32.
- Feng T, Lu Z, Wei Y, Meng J, Lin CP, et al. PDGFRb+ mesenchymal cells, but not NG2+mural cells, contribute to cardiac fat. iang Z. Cell rep. 2021; 34: 108697.
- Moore-Morris T, Cattaneo P, Puceat M, Evans SM. Origins of cardiac fibroblasts. J Mol Cell Cardiol. 2016; 91: 1-5.
- Ratajska A, Czarnowska E, Kolodzinska A, Kluzek W, Lesniak W. Vasculogenesis of the embryonic heart: origin of blood island-like structures. Anat Rec A Discov Mol Cell Evol Biol. 2006; 288: 223-32.
- Takeichi M, Nimura K, Mori M, Nakagami H, Kaneda Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLOS ONE. 2013; 8: e57829.
- Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol. 2012; 366: 111-24.
- Liu K, Yu W, Tang M, Tang J, Liu X, Liu Q, et al. A dual genetic tracing system identifies diverse and dynamic origins of cardiac valve mesenchyme. Development. 2018; 145: dev167775.
- Wolters R, Deepe R, Drummond J, Harvey AB, Hiriart E, Lockhart MM, et al. Role of the epicardium in the development of the atrioventricular valves and its relevance to the pathogenesis of myxomatous valve disease. J Cardiovasc Dev Dis. 2021; 8: 54.
- Cano E, Carmona R, Ruiz-Villalba A, Rojas A, Chau YY, Wagner KD, et al. Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio-venous connections. Proc Natl Acad Sci USA. 2016; 113: 656-61.
- Zhou B, Pu WT. Genetic Cre-loxP assessment of epicardial cell fate using Wt1-driven Cre alleles. Circ Res. 2012; 111: e276-80.
- Yang DY, Song HY, Zhang HQ, Huang XY, Guan XQ. Preliminary study of ALK3 downstream genes related to ventricular septum defect]. Sheng Wu Gong Cheng Xue Bao. 2003; 19: 267-71.
- Bargehr J. Bargehr J, Ong LP, Colzani M, Davaapil H, Hofsteen P, et al. Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nat Biotechnol. 2019; 37: 895-906.
- Del Campo CV, Liaw NY, Gunadasa-Rohling M, Matthaei M, Braga L, Kennedy T, et al. Regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer. Cardiovasc Res. 2022; 118: 597-611.
- Martínez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet. 2010; 42: 89-93.